Sample the Medication Data Recorded in the UK Biobank Was Collected Through a Verbal Interview with a Trained Nurse
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Appendix A: Potentially Inappropriate Prescriptions (Pips) for Older People (Modified from ‘STOPP/START 2’ O’Mahony Et Al 2014)
Appendix A: Potentially Inappropriate Prescriptions (PIPs) for older people (modified from ‘STOPP/START 2’ O’Mahony et al 2014) Consider holding (or deprescribing - consult with patient): 1. Any drug prescribed without an evidence-based clinical indication 2. Any drug prescribed beyond the recommended duration, where well-defined 3. Any duplicate drug class (optimise monotherapy) Avoid hazardous combinations e.g.: 1. The Triple Whammy: NSAID + ACE/ARB + diuretic in all ≥ 65 year olds (NHS Scotland 2015) 2. Sick Day Rules drugs: Metformin or ACEi/ARB or a diuretic or NSAID in ≥ 65 year olds presenting with dehydration and/or acute kidney injury (AKI) (NHS Scotland 2015) 3. Anticholinergic Burden (ACB): Any additional medicine with anticholinergic properties when already on an Anticholinergic/antimuscarinic (listed overleaf) in > 65 year olds (risk of falls, increased anticholinergic toxicity: confusion, agitation, acute glaucoma, urinary retention, constipation). The following are known to contribute to the ACB: Amantadine Antidepressants, tricyclic: Amitriptyline, Clomipramine, Dosulepin, Doxepin, Imipramine, Nortriptyline, Trimipramine and SSRIs: Fluoxetine, Paroxetine Antihistamines, first generation (sedating): Clemastine, Chlorphenamine, Cyproheptadine, Diphenhydramine/-hydrinate, Hydroxyzine, Promethazine; also Cetirizine, Loratidine Antipsychotics: especially Clozapine, Fluphenazine, Haloperidol, Olanzepine, and phenothiazines e.g. Prochlorperazine, Trifluoperazine Baclofen Carbamazepine Disopyramide Loperamide Oxcarbazepine Pethidine -

Chlorphenamine Maleate)
Package leaflet: Information for the patient Chlorphenamine 10 mg/ml Solution for Injection (Chlorphenamine Maleate) Read all of this leaflet carefully before you start taking this medicine because it contains important information for you. − Keep this leaflet. You may need to read it again. − If you have any further questions, ask your doctor or nurse. − If you get any side effects, talk to your doctor or nurse. This includes any possible side effects not listed in this leaflet. See section 4. What is in this leaflet: 1. What Chlorphenamine is and what it is used for 2. What you need to know before Chlorphenamine is given 3. How Chlorphenamine is given 4. Possible side effects 5. How to store Chlorphenamine 6. Contents of the pack and other information 1. What Chlorphenamine is and what it is used for Chlorphenamine 10 mg/ml Solution for Injection contains the active ingredient chlorphenamine maleate which is an antihistamine. Chlorphenamine is indicated in adults and children (aged 1 month to 18 years) for the treatment of acute allergic reactions. These medicines inhibit the release of histamine into the body that occurs during an allergic reaction. This product relieves some of the main symptoms of a severe allergic reaction. 2. What you need to know before Chlorphenamine is given You MUST NOT be given Chlorphenamine: if you are allergic to chlorphenamine maleate or any of the other ingredients of this medicine (listed in section 6) if you have had monoamine oxidase inhibitor (MAOI) antidepressive treatment within the past 14 days. Warnings and precautions Talk to your doctor or nurse before you are given this medicine if you: are being treated for an overactive thyroid or enlarged prostate gland have epilepsy, raised blood pressure within the eye or glaucoma, very high blood pressure, heart, liver, asthma or other chest diseases. -

Prescribing Trends of Antihistamines in the Outpatient Setting in Al-Kharj
Prescribing Trends of Antihistamines in the Outpatient Setting in Al-Kharj Nehad J. Ahmed1*, Menshawy A. Menshawy2 1Department of Clinical Pharmacy, College of Pharmacy, Prince Sattam Bin Abdulaziz University, Al-Kharj, Saudi Arabia, 2Department of Medicinal chemistry, College of Pharmacy, Prince Sattam Bin Abdulaziz University, Al-Kharj, Saudi Arabia Abstract Aim: This study aims to illustrate the prescribing trends of antihistamines in the outpatient setting in Al-Kharj. Materials and Methods: This is a retrospective study that included the evaluation of antihistamines in the outpatient setting in a public hospital in Al-Kharj. The data were collected from the pharmacy-based computer system. Results: The total number of prescriptions that included antihistamines was 799. Most of the prescribed ORIGINAL ARTICLE ARTICLE ORIGINAL antihistamines were first-generation sedating antihistamines (chlorphenamine and diphenhydramine) (66.33%). About 63.20% of the prescribed antihistamines included chlorpheniramine followed by cetirizine (19.27%) and loratadine (14.39%). Conclusion: Antihistamines were prescribed commonly in the outpatient setting mainly first-generation sedating antihistamines. It is recommended to increase the awareness of health- care providers about the efficacy and the side effects of antihistamines and to encourage them to use these agents wisely. Key words: Antihistamines, outpatient, prescribing, trends INTRODUCTION In addition, antihistamines have been classified as sedating antihistamines (first-generation antihistamines) and non- ntihistamines are used in the sedating antihistamines (second-generation antihistamines).[4] management of allergic conditions. Sedating antihistamines include chlorphenamine, clemastine, They are useful for treating the itching hydroxyzine, alimemazine, cyproheptadine, promethazine, A [1] and ketotifen.[4] Non-sedating antihistamines include that results from the release of histamine. -

(12) Patent Application Publication (10) Pub. No.: US 2006/0110428A1 De Juan Et Al
US 200601 10428A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2006/0110428A1 de Juan et al. (43) Pub. Date: May 25, 2006 (54) METHODS AND DEVICES FOR THE Publication Classification TREATMENT OF OCULAR CONDITIONS (51) Int. Cl. (76) Inventors: Eugene de Juan, LaCanada, CA (US); A6F 2/00 (2006.01) Signe E. Varner, Los Angeles, CA (52) U.S. Cl. .............................................................. 424/427 (US); Laurie R. Lawin, New Brighton, MN (US) (57) ABSTRACT Correspondence Address: Featured is a method for instilling one or more bioactive SCOTT PRIBNOW agents into ocular tissue within an eye of a patient for the Kagan Binder, PLLC treatment of an ocular condition, the method comprising Suite 200 concurrently using at least two of the following bioactive 221 Main Street North agent delivery methods (A)-(C): Stillwater, MN 55082 (US) (A) implanting a Sustained release delivery device com (21) Appl. No.: 11/175,850 prising one or more bioactive agents in a posterior region of the eye so that it delivers the one or more (22) Filed: Jul. 5, 2005 bioactive agents into the vitreous humor of the eye; (B) instilling (e.g., injecting or implanting) one or more Related U.S. Application Data bioactive agents Subretinally; and (60) Provisional application No. 60/585,236, filed on Jul. (C) instilling (e.g., injecting or delivering by ocular ion 2, 2004. Provisional application No. 60/669,701, filed tophoresis) one or more bioactive agents into the Vit on Apr. 8, 2005. reous humor of the eye. Patent Application Publication May 25, 2006 Sheet 1 of 22 US 2006/0110428A1 R 2 2 C.6 Fig. -

The Effects of Antipsychotic Treatment on Metabolic Function: a Systematic Review and Network Meta-Analysis
The effects of antipsychotic treatment on metabolic function: a systematic review and network meta-analysis Toby Pillinger, Robert McCutcheon, Luke Vano, Katherine Beck, Guy Hindley, Atheeshaan Arumuham, Yuya Mizuno, Sridhar Natesan, Orestis Efthimiou, Andrea Cipriani, Oliver Howes ****PROTOCOL**** Review questions 1. What is the magnitude of metabolic dysregulation (defined as alterations in fasting glucose, total cholesterol, low density lipoprotein (LDL) cholesterol, high density lipoprotein (HDL) cholesterol, and triglyceride levels) and alterations in body weight and body mass index associated with short-term (‘acute’) antipsychotic treatment in individuals with schizophrenia? 2. Does baseline physiology (e.g. body weight) and demographics (e.g. age) of patients predict magnitude of antipsychotic-associated metabolic dysregulation? 3. Are alterations in metabolic parameters over time associated with alterations in degree of psychopathology? 1 Searches We plan to search EMBASE, PsycINFO, and MEDLINE from inception using the following terms: 1 (Acepromazine or Acetophenazine or Amisulpride or Aripiprazole or Asenapine or Benperidol or Blonanserin or Bromperidol or Butaperazine or Carpipramine or Chlorproethazine or Chlorpromazine or Chlorprothixene or Clocapramine or Clopenthixol or Clopentixol or Clothiapine or Clotiapine or Clozapine or Cyamemazine or Cyamepromazine or Dixyrazine or Droperidol or Fluanisone or Flupehenazine or Flupenthixol or Flupentixol or Fluphenazine or Fluspirilen or Fluspirilene or Haloperidol or Iloperidone -

Your Child's Emergency Allergy Pack with Antihistamine
Your Child’s Emergency Allergy Pack with Antihistamine Patient information Paediatric Department Watford General Hospital Hemel Hempstead Hospital If you need this leaflet in another language, large print, Braille or audio version, please call 01923 217 187 or email [email protected] Author Dr Ashley Reece Department Paediatrics Ratified Date / Review Date Feb 2021 / Feb 2024 Version Number / ID Number 40-1104-V1 Why does my child I need an Allergy Action Pack? An allergy action pan is a kit contianing evrything you need in case of an allergic reaction. Is my/ my child’s allergy severe? There is no such thing as a mild or severe allergy as reactions are always unpredicatble. However we do classify reactions as mild, moderate or severe. While it follows that a severe reaction makes the anxiety about the severity of the next reaction high, we cannot predict the severity of subsequent reactions. However there are some foods and some allergies which are assessed as having very low risk of a significant reaction. The way to manage your child with their allergy is to ensure you have the best possible safety net in case of an unforeseen reaction. Your doctor will give you a specific allergy plan (like the one to the right) which you should also give to your child’s school. Generally allergy is mananged by: You MUST avoid any foods which you know your child is allergic to. Avoidance Take care with labels and risk assess any new foods and when eating out in a restaurant or from a take-away. -

A Nested Case–Control Study
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Golestan University of Medical Sciences Repository Schizophrenia Research 150 (2013) 416–420 Contents lists available at ScienceDirect Schizophrenia Research journal homepage: www.elsevier.com/locate/schres Medication and suicide risk in schizophrenia: A nested case–control study Johan Reutfors a,⁎, Shahram Bahmanyar a,b, Erik G. Jönsson c, Lena Brandt a, Robert Bodén a,d, Anders Ekbom a, Urban Ösby e,f a Centre for Pharmacoepidemiology (CPE), Department of Medicine Solna, Karolinska Institutet, Stockholm, Sweden b Faculty of Medicine, Golestan University of Medical Sciences, Gorgan, Iran c Department of Clinical Neuroscience, Karolinska Institutet, Stockholm, Sweden d Department of Neuroscience, Psychiatry, Uppsala University, Uppsala, Sweden e Department of Neurobiology, Care Sciences, and Society, Centre of Family Medicine (CeFAM), Karolinska Institutet, Stockholm, Sweden f Department of Psychiatry, Tiohundra AB, Norrtälje, Sweden article info abstract Article history: Introduction: Patients with schizophrenia are at increased risk of suicide, but data from controlled studies of Received 16 June 2013 pharmacotherapy in relation to suicide risk is limited. Received in revised form 28 August 2013 Aim: To explore suicide risk in schizophrenia in relation to medication with antipsychotics, antidepressants, Accepted 9 September 2013 and lithium. Available online 2 October 2013 Methods: Of all patients with a first clinical discharge diagnosis of schizophrenia or schizoaffective disorder in Stockholm County between 1984 and 2000 (n = 4000), patients who died by suicide within five years Keywords: from diagnosis were defined as cases (n = 84; 54% male). Individually matched controls were identified Schizophrenia Suicide from the same population. -

Antipsychotics and the Risk of Sudden Cardiac Death
ORIGINAL INVESTIGATION Antipsychotics and the Risk of Sudden Cardiac Death Sabine M. J. M. Straus, MD; Gyse`le S. Bleumink, MD; Jeanne P. Dieleman, PhD; Johan van der Lei, MD, PhD; Geert W. ‘t Jong, PhD; J. Herre Kingma, MD, PhD; Miriam C. J. M. Sturkenboom, PhD; Bruno H. C. Stricker, PhD Background: Antipsychotics have been associated with Results: The study population comprised 554 cases of prolongation of the corrected QT interval and sudden car- sudden cardiac death. Current use of antipsychotics was diac death. Only a few epidemiological studies have in- associated with a 3-fold increase in risk of sudden car- vestigated this association. We performed a case- diac death. The risk of sudden cardiac death was high- control study to investigate the association between use est among those using butyrophenone antipsychotics, of antipsychotics and sudden cardiac death in a well- those with a defined daily dose equivalent of more than defined community-dwelling population. 0.5 and short-term (Յ90 days) users. The association with current antipsychotic use was higher for witnessed cases Methods: We performed a population-based case-control (n=334) than for unwitnessed cases. study in the Integrated Primary Care Information (IPCI) project, a longitudinal observational database with com- Conclusions: Current use of antipsychotics in a gen- plete medical records from 150 general practitioners. All eral population is associated with an increased risk of sud- instances of death between January 1, 1995, and April 1, den cardiac death, even at a low dose and for indica- 2001, were reviewed. Sudden cardiac death was classified tions other than schizophrenia. -

Importance of the Newer Generations Of
Psychiatria Danubina, 2013; Vol. 25, No. 3, pp 329-333 Conference paper © Medicinska naklada - Zagreb, Croatia IMPORTANCE OF THE NEWER GENERATIONS OF ANTIPSYCHOTICS IN REDUCING SCHIZOPHRENIA HOSPITALIZATION RATES Vlado Jukić, Aleksandar Savić & Miroslav Herceg University Psychiatric Hospital Vrapče, Zagreb, Croatia SUMMARY Throughout history, given the lack of understanding of schizophrenia and lack of effective treatment options, patients were often committed to asylums and later psychiatric institutions, often for prolonged periods of time. First antipsychotic medications helped to bring about changes in approach to these patients and facilitated deinstitutionalization, and discovery of new drugs with differing side-effects profiles introduced new options in treating schizophrenia patients. Data on hospitalization of patients in University Psychiatric Hospital Vrapče from the mid-1990s, as well as data on national level, suggests a trend of drop in hospitalization of schizophrenia patients. At the same time, that period saw significant increase in a number of available newer-generations antipsychotics and the rise in their use compared to first-generation one. Although far from being the only contributing factor, seem to play an important role in continuing the trend of reducing hospitalization rates for schizophrenia patients that started with first antipsychotics. Newer antipsychotics with a more tolerable side-effects profile promote better compliance and further reduce rate of relapse and hospitalizations. No less important is the contribution of newer antipsychotics with new receptor profiles to the personalized psychopharmacotherapy approach that is in tune with emerging conceptualizations of schizophrenia as a complex syndrome with a number of separate symptom domains, whose specific combinations produce specific individual clinical presentation and in turn ask for a specific individual approach to the patient. -
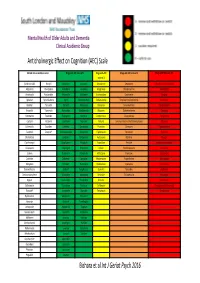
AEC Traffic Light System
Mental Health of Older Adults and Dementia Clinical Academic Group Anticholinergic Effect on Cognition (AEC) Scale Limited data so unable to score Drugs with AEC score of 0 Drugs with AEC Drugs with AEC score of 2 Drugs with AEC score of 3 score of 1 Alendronic Acid Ramipril Alprazolam Lovastatin Amiodarone Amantadine Alimemazine (trimeprazine) Allopurinol Rivaroxaban Amlodipine Lurasidone Aripiprazole Chlorphenamine Amitriptyline Anastrozole Rosuvastatin Amoxycillin Meloxicam Bromocriptine Desipramine Atropine Apixaban Spironolactone Aspirin Metoclopramide Carbamazepine Dicycloverine (dicyclomine) Benztropine Baclofen Tamoxifen Atenolol Metoprolol Citalopram Dimenhydrinate Chlorpromazine Bisoprolol Topiramate Atorvastatin Moclobemide Diazepam Diphenhydramine Clemastine Bumetanide Tizanidine Buproprion Morphine Domperidone Disopyramide Clomipramine Captopril Verapamil Cepahlexin Naproxen Fentanyl Levomepromazine (methotrimeprazine) Clozapine Carbimazole Zopiclone Cetirizine Omeprazole Fluoxetine Olanzapine Cyproheptadine Carvedilol Zotepine* Chlordiazepoxide Paracetamol Fluphenazine Paroxetine Dothiepin Chlortalidone Cimetidine Pantoprazole Hydroxyzine Pethidine Doxepin Clarithromycin Ciprofloxacin Pravastatin Iloperidone Pimozide Hyoscine hydrobromide Clonazepam Clopidogrel Propranolol Lithium Prochlorperazine Imipramine Codeine Darifenacin Rabeprazole Mirtazapine Promazine Lofepramine Colchicine Diclofenac Ranitidine Perphenazine Propantheline Nortriptyline Dabigatran Diltiazem Risperidone Prednisolone Quetiapine Orphenadrine Dexamethasone -

Summary of Product Characteristics
SUMMARY OF PRODUCT CHARACTERISTICS 1 1. NAME OF THE MEDICINAL PRODUCT (Invented name), film-coated tablet 2. QUALITATIVE AND QUANTITATIVE COMPOSITION Paracetamol..............................................................................................................................500.00 mg Chlorphenamine maleate..............................................................................................................4.00 mg For one film-coated tablet Excipients with known effect: Carmoisine (E122) For the full list of excipients, see section 6.1. 3. PHARMACEUTICAL FORM Oblong purple film-coated tablet. 4. CLINICAL PARTICULARS 4.1 Therapeutic indications This medicinal product is indicated for treatment, during the course of colds, rhinitis, rhinopharyngitis and flu-like conditions in adults and children aged over 15 years: of clear nasal discharge and watering of the eyes, of sneezing, of headaches and/or fever. 4.2 Posology and method of administration Posology Restricted to adults and children over the age of 15 years. Weight Dose per Administration Maximum daily dose (age) administration interval Adults and 1 tablet 4 hours 4 tablets children >50 kg i.e., i.e., (>15 years) 500 mg of paracetamol 2,000 mg of paracetamol 4 mg of chlorphenamine 16 mg of chlorphenamine Do not exceed the maximum posology of 4 tablets per 24 hours. Patients with renal failure In the event of renal failure and unless otherwise medically advised, it is recommended that the dose be reduced and that the minimum interval between the 2 doses be increased, based on the following table: 2 Creatinine Administration interval clearance ≥50 ml/min 4 hours 10–50 ml/min 6 hours <10 ml/min 8 hours The total dose of paracetamol should not exceed 3 g/day. Patients with hepatic failure In patients with active or compensated chronic hepatic disease, particularly those with hepatocellular insufficiency, chronic alcoholism, chronic malnutrition (low reserves of hepatic glutathione), and dehydration, the dose of paracetamol should not exceed 3 g/day. -

Drug and Medication Classification Schedule
KENTUCKY HORSE RACING COMMISSION UNIFORM DRUG, MEDICATION, AND SUBSTANCE CLASSIFICATION SCHEDULE KHRC 8-020-1 (11/2018) Class A drugs, medications, and substances are those (1) that have the highest potential to influence performance in the equine athlete, regardless of their approval by the United States Food and Drug Administration, or (2) that lack approval by the United States Food and Drug Administration but have pharmacologic effects similar to certain Class B drugs, medications, or substances that are approved by the United States Food and Drug Administration. Acecarbromal Bolasterone Cimaterol Divalproex Fluanisone Acetophenazine Boldione Citalopram Dixyrazine Fludiazepam Adinazolam Brimondine Cllibucaine Donepezil Flunitrazepam Alcuronium Bromazepam Clobazam Dopamine Fluopromazine Alfentanil Bromfenac Clocapramine Doxacurium Fluoresone Almotriptan Bromisovalum Clomethiazole Doxapram Fluoxetine Alphaprodine Bromocriptine Clomipramine Doxazosin Flupenthixol Alpidem Bromperidol Clonazepam Doxefazepam Flupirtine Alprazolam Brotizolam Clorazepate Doxepin Flurazepam Alprenolol Bufexamac Clormecaine Droperidol Fluspirilene Althesin Bupivacaine Clostebol Duloxetine Flutoprazepam Aminorex Buprenorphine Clothiapine Eletriptan Fluvoxamine Amisulpride Buspirone Clotiazepam Enalapril Formebolone Amitriptyline Bupropion Cloxazolam Enciprazine Fosinopril Amobarbital Butabartital Clozapine Endorphins Furzabol Amoxapine Butacaine Cobratoxin Enkephalins Galantamine Amperozide Butalbital Cocaine Ephedrine Gallamine Amphetamine Butanilicaine Codeine