Cross-Species Transcriptomic Analysis Elucidates Constitutive Aryl
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma
Anatomy and Pathology Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma Sarah L. Lake,1 Sarah E. Coupland,1 Azzam F. G. Taktak,2 and Bertil E. Damato3 PURPOSE. To detect deletions and loss of heterozygosity of disease is fatal in 92% of patients within 2 years of diagnosis. chromosome 3 in a rare subset of fatal, disomy 3 uveal mela- Clinical and histopathologic risk factors for UM metastasis noma (UM), undetectable by fluorescence in situ hybridization include large basal tumor diameter (LBD), ciliary body involve- (FISH). ment, epithelioid cytomorphology, extracellular matrix peri- ϩ ETHODS odic acid-Schiff-positive (PAS ) loops, and high mitotic M . Multiplex ligation-dependent probe amplification 3,4 5 (MLPA) with the P027 UM assay was performed on formalin- count. Prescher et al. showed that a nonrandom genetic fixed, paraffin-embedded (FFPE) whole tumor sections from 19 change, monosomy 3, correlates strongly with metastatic death, and the correlation has since been confirmed by several disomy 3 metastasizing UMs. Whole-genome microarray analy- 3,6–10 ses using a single-nucleotide polymorphism microarray (aSNP) groups. Consequently, fluorescence in situ hybridization were performed on frozen tissue samples from four fatal dis- (FISH) detection of chromosome 3 using a centromeric probe omy 3 metastasizing UMs and three disomy 3 tumors with Ͼ5 became routine practice for UM prognostication; however, 5% years’ metastasis-free survival. to 20% of disomy 3 UM patients unexpectedly develop metas- tases.11 Attempts have therefore been made to identify the RESULTS. Two metastasizing UMs that had been classified as minimal region(s) of deletion on chromosome 3.12–15 Despite disomy 3 by FISH analysis of a small tumor sample were found these studies, little progress has been made in defining the key on MLPA analysis to show monosomy 3. -

Fine–Mapping Identifies NAD–ME1 As a Candidate Underlying a Major
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.07.285429; this version posted September 9, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Fine–mapping identifies NAD–ME1 as a candidate underlying 2 a major locus controlling temporal variation in primary and 3 specialized metabolism in Arabidopsis 4 5 Marta Francisco1, Daniel J. Kliebenstein2,3, Víctor M. Rodríguez1, Pilar Soengas1, 6 Rosaura Abilleira1, María E. Cartea1 7 1Misión Biológica de Galicia, (MBG-CSIC), P.O. Box 28, 36080, Pontevedra, Spain 8 2Department of Plant Sciences, University of California at Davis, Davis, CA 95616, 9 USA. 10 3DynaMo Center of Excellence, University of Copenhagen, Thorvaldsensvej 40, DK- 11 1871 Frederiksberg C, Denmark. 12 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.07.285429; this version posted September 9, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 13 Summary 14 Plant metabolism is modulated by a complex interplay between internal signals and 15 external cues. A major goal of all quantitative metabolomic studies is to clone the 16 underlying genes to understand the mechanistic basis of this variation. Using fine-scale 17 genetic mapping, in this work we report the identification and initial characterization of 18 NAD-DEPENDENT MALIC ENZYME 1 (NAD-ME1) as the candidate gene underlying 19 the pleiotropic network Met.II.15 QTL controlling variation in plant metabolism and 20 circadian clock outputs in the Bay × Sha Arabidopsis population. -

Profiling Cellular Processes in Adipose Tissue During Weight Loss Using Time Series Gene Expression
G C A T T A C G G C A T genes Article Profiling Cellular Processes in Adipose Tissue during Weight Loss Using Time Series Gene Expression Samar H. K. Tareen 1,* , Michiel E. Adriaens 1,* , Ilja C. W. Arts 1,2, Theo M. de Kok 1,3, Roel G. Vink 4, Nadia J. T. Roumans 4, Marleen A. van Baak 4, Edwin C. M. Mariman 4, Chris T. Evelo 1,5,* and Martina Kutmon 1,5,* 1 Maastricht Centre for Systems Biology (MaCSBio), Maastricht University, 6211ER Maastricht, The Netherlands; [email protected] (I.C.W.A.); [email protected] (T.M.d.K.) 2 Department of Epidemiology, CARIM School for Cardiovascular Diseases, Maastricht University, 6211ER Maastricht, The Netherlands 3 Department of Toxicogenomics, GROW School of Oncology and Developmental Biology, Maastricht University, 6211ER Maastricht, The Netherlands 4 Department of Human Biology, NUTRIM Research School, Maastricht University, 6211ER Maastricht, The Netherlands; [email protected] (R.G.V.); [email protected] (N.J.T.R.); [email protected] (M.A.v.B.); [email protected] (E.C.M.M.) 5 Department of Bioinformatics—BiGCaT, NUTRIM Research School, Maastricht University, 6211ER Maastricht, The Netherlands * Correspondence: [email protected] (S.H.K.T.); [email protected] (M.E.A.); [email protected] (C.T.E.); [email protected] (M.K.) Received: 28 September 2018; Accepted: 22 October 2018; Published: 29 October 2018 Abstract: Obesity is a global epidemic identified as a major risk factor for multiple chronic diseases and, consequently, diet-induced weight loss is used to counter obesity. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -

Mouse Me1 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Me1 Knockout Project (CRISPR/Cas9) Objective: To create a Me1 knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Me1 gene (NCBI Reference Sequence: NM_001198933 ; Ensembl: ENSMUSG00000032418 ) is located on Mouse chromosome 9. 14 exons are identified, with the ATG start codon in exon 1 and the TAA stop codon in exon 14 (Transcript: ENSMUST00000185374). Exon 2~3 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mice homozygous for a spontaneous allele exhibit decreased body weight on a medium fat diet, altered cytoplasmic malic enzyme activity, and a male-specific reduction in food intake on a high fat diet. Exon 2 starts from about 1.15% of the coding region. Exon 2~3 covers 17.15% of the coding region. The size of effective KO region: ~2089 bp. The KO region does not have any other known gene. Page 1 of 9 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 2 3 14 Legends Exon of mouse Me1 Knockout region Page 2 of 9 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section upstream of Exon 2 is aligned with itself to determine if there are tandem repeats. No significant tandem repeat is found in the dot plot matrix. So this region is suitable for PCR screening or sequencing analysis. -

Genetic Commonalities in Gynecologic Cancers Using Publicly Available Genome-Wide Association Study Summary Results: an Exploratory Meta-Analysis
Title Page Genetic Commonalities in Gynecologic Cancers Using Publicly Available Genome-Wide Association Study Summary Results: An Exploratory Meta-Analysis by Mark Francis Vater BS Biology, Northern Kentucky University, 2014 Submitted to the Graduate Faculty of the Graduate School of Public Health in partial fulfillment of the requirements for the degree of Master of Science University of Pittsburgh 2021 Committee Membership Page UNIVERSITY OF PITTSBURGH GRADUATE SCHOOL OF PUBLIC HEALTH This thesis was presented by Mark Francis Vater It was defended on August 4, 2021 and approved by Jeanine Buchanich, Associate Professor, Department of Biostatistics Ada Youk, Associate Professor, Department of Biostatistics John Shaffer, Assistant Professor, Department of Human Genetics Thesis Advisor: Jenna Carlson, Assistant Professor, Department of Biostatistics ii Copyright © by Mark Vater 2021 iii Abstract Genetic Commonalities in Gynecologic Cancers Using Publicly Available Genome-Wide Association Study Summary Results: An Exploratory Meta-Analysis Mark Francis Vater, MS University of Pittsburgh, 2021 Abstract Public Health Significance: Gynecologic cancers are responsible for millions of deaths worldwide every year. The goal of this research is to further scientific understanding of such cancers, potentially leading to improved diagnosis and treatment. The public health significance of this project is to uncover potential genetic underpinnings of gynecologic cancers, aiding in efforts to reduce the mortality rate of gynecologic cancers, which is imperative in protecting the health of susceptible persons across the globe. Gynecologic cancers are those which arise in the female reproductive system, chiefly, those of the ovaries, cervix, vulva, endometrium, and vagina. These conditions present a serious threat to the health of susceptible persons, leading to nearly three million deaths worldwide each year. -

Dual Specificity Phosphatases from Molecular Mechanisms to Biological Function
International Journal of Molecular Sciences Dual Specificity Phosphatases From Molecular Mechanisms to Biological Function Edited by Rafael Pulido and Roland Lang Printed Edition of the Special Issue Published in International Journal of Molecular Sciences www.mdpi.com/journal/ijms Dual Specificity Phosphatases Dual Specificity Phosphatases From Molecular Mechanisms to Biological Function Special Issue Editors Rafael Pulido Roland Lang MDPI • Basel • Beijing • Wuhan • Barcelona • Belgrade Special Issue Editors Rafael Pulido Roland Lang Biocruces Health Research Institute University Hospital Erlangen Spain Germany Editorial Office MDPI St. Alban-Anlage 66 4052 Basel, Switzerland This is a reprint of articles from the Special Issue published online in the open access journal International Journal of Molecular Sciences (ISSN 1422-0067) from 2018 to 2019 (available at: https: //www.mdpi.com/journal/ijms/special issues/DUSPs). For citation purposes, cite each article independently as indicated on the article page online and as indicated below: LastName, A.A.; LastName, B.B.; LastName, C.C. Article Title. Journal Name Year, Article Number, Page Range. ISBN 978-3-03921-688-8 (Pbk) ISBN 978-3-03921-689-5 (PDF) c 2019 by the authors. Articles in this book are Open Access and distributed under the Creative Commons Attribution (CC BY) license, which allows users to download, copy and build upon published articles, as long as the author and publisher are properly credited, which ensures maximum dissemination and a wider impact of our publications. The book as a whole is distributed by MDPI under the terms and conditions of the Creative Commons license CC BY-NC-ND. Contents About the Special Issue Editors .................................... -

Dissecting the Genetic Etiology of Lupus at ETS1 Locus
Dissecting the Genetic Etiology of Lupus at ETS1 Locus A dissertation submitted to the Graduate School of the University of Cincinnati in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Immunobiology of the College of Medicine 2017 by Xiaoming Lu B.S. Sun Yat-sen University, P.R. China June 2011 Dissertation Committee: John B. Harley, MD, PhD Harinder Singh, PhD Leah C. Kottyan, PhD Matthew T. Weirauch, PhD Kasper Hoebe, PhD Lili Ding, PhD i Abstract Systemic lupus erythematosus (SLE) is a complex autoimmune disease with strong evidence for genetics factor involvement. Genome-wide association studies have identified 84 risk loci associated with SLE. However, the specific genotype-dependent (allelic) molecular mechanisms connecting these lupus-genetic risk loci to immunological dysregulation are mostly still unidentified. ~ 90% of these loci contain variants that are non-coding, and are thus likely to act by impacting subtle, comparatively hard to predict mechanisms controlling gene expression. Here, we developed a strategic approach to prioritize non-coding variants, and screen them for their function. This approach involves computational prioritization using functional genomic databases followed by experimental analysis of differential binding of transcription factors (TFs) to risk and non-risk alleles. For both electrophoretic mobility shift assay (EMSA) and DNA affinity precipitation assay (DAPA) analysis of genetic variants, a synthetic DNA oligonucleotide (oligo) is used to identify factors in the nuclear lysate of disease or phenotype-relevant cells. This strategic approach was then used for investigating SLE association at ETS1 locus. Genetic variants at chromosomal region 11q23.3, near the gene ETS1, have been associated with systemic lupus erythematosus (SLE), or lupus, in independent cohorts of Asian ancestry. -
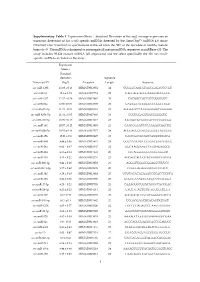
Supplementary Table 1. Expression
Supplementary Table 1. Expression (Mean Standard Deviation of the log2 average expression or transcript detection) of Sus scrofa specific miRNAs detected by the GeneChip™ miRNA 4.0 Array (ThermoFisher Scientific) in spermatozoa retrieved from the SRF of the ejaculate of healthy mature boars (n=3). The miRNA is designed to interrogate all mature miRNA sequences in miRBase v20. The array includes 30.424 mature miRNA (all organisms) and we select specifically the 326 Sus scrofa- specific miRNAs included in the array. Expression Mean ± Standard Deviation Sequence Transcript ID (log2) Accession Length Sequence ssc-miR-1285 13.98 ± 0.13 MIMAT0013954 24 CUGGGCAACAUAGCGAGACCCCGU ssc-miR-16 12.6 ± 0.74 MIMAT0007754 22 UAGCAGCACGUAAAUAUUGGCG ssc-miR-4332 12.32 ± 0.29 MIMAT0017962 20 CACGGCCGCCGCCGGGCGCC ssc-miR-92a 12.06 ± 0.09 MIMAT0013908 22 UAUUGCACUUGUCCCGGCCUGU ssc-miR-671-5p 11.73 ± 0.54 MIMAT0025381 24 AGGAAGCCCUGGAGGGGCUGGAGG ssc-miR-4334-5p 11.31 ± 0.05 MIMAT0017966 19 CCCUGGAGUGACGGGGGUG ssc-miR-425-5p 10.99 ± 0.15 MIMAT0013917 23 AAUGACACGAUCACUCCCGUUGA ssc-miR-191 10.57 ± 0.22 MIMAT0013876 23 CAACGGAAUCCCAAAAGCAGCUG ssc-miR-92b-5p 10.53 ± 0.18 MIMAT0017377 24 AGGGACGGGACGCGGUGCAGUGUU ssc-miR-15b 10.01 ± 0.9 MIMAT0002125 22 UAGCAGCACAUCAUGGUUUACA ssc-miR-30d 9.89 ± 0.36 MIMAT0013871 24 UGUAAACAUCCCCGACUGGAAGCU ssc-miR-26a 9.62 ± 0.47 MIMAT0002135 22 UUCAAGUAAUCCAGGAUAGGCU ssc-miR-484 9.55 ± 0.14 MIMAT0017974 20 CCCAGGGGGCGACCCAGGCU ssc-miR-103 9.53 ± 0.22 MIMAT0002154 23 AGCAGCAUUGUACAGGGCUAUGA ssc-miR-296-3p 9.41 ± 0.26 MIMAT0022958 -

Mouse Me1 Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Me1 Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Me1 conditional knockout Mouse model (C57BL/6N) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Me1 gene (NCBI Reference Sequence: NM_001198933 ; Ensembl: ENSMUSG00000032418 ) is located on Mouse chromosome 9. 14 exons are identified, with the ATG start codon in exon 1 and the TAA stop codon in exon 14 (Transcript: ENSMUST00000185374). Exon 2 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Me1 gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP23-56G17 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mice homozygous for a spontaneous allele exhibit decreased body weight on a medium fat diet, altered cytoplasmic malic enzyme activity, and a male-specific reduction in food intake on a high fat diet. Exon 2 starts from about 1.15% of the coding region. The knockout of Exon 2 will result in frameshift of the gene. The size of intron 1 for 5'-loxP site insertion: 7005 bp, and the size of intron 2 for 3'-loxP site insertion: 1805 bp. The size of effective cKO region: ~634 bp. The cKO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele gRNA region 5' gRNA region 3' 1 2 3 14 Targeting vector Targeted allele Constitutive KO allele (After Cre recombination) Legends Exon of mouse Me1 Homology arm cKO region loxP site Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Mouse Tiparp Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Tiparp Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Tiparp conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Tiparp gene (NCBI Reference Sequence: NM_178892 ; Ensembl: ENSMUSG00000034640 ) is located on Mouse chromosome 3. 6 exons are identified, with the ATG start codon in exon 2 and the TGA stop codon in exon 6 (Transcript: ENSMUST00000047906). Exon 2 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Tiparp gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP23-371A15 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mice homozygous for a gene trapped allele exhibit postnatal lethality, skeletal and craniofacial defects, kidney defects and embryonic hemorrhaging. Exon 2 starts from about 0.1% of the coding region. The knockout of Exon 2 will result in frameshift of the gene. The size of intron 1 for 5'-loxP site insertion: 2591 bp, and the size of intron 2 for 3'-loxP site insertion: 14022 bp. The size of effective cKO region: ~1420 bp. The cKO region does not have any other known gene. Page 1 of 7 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 2 6 Targeting vector Targeted allele Constitutive KO allele (After Cre recombination) Legends Exon of mouse Tiparp Homology arm cKO region loxP site Page 2 of 7 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Supplemental Materials and Methods Page
1 2 Supplementary Information for 3 4 5 Vertebrate adaptive radiation is assembled from an ancient and disjunct spatiotemporal 6 landscape 7 8 Emilie J. Richards, Joseph A. McGirr, Jeremy R. Wang, Michelle E. St. John, Jelmer W. 9 Poelstra, Maria J. Solano, Delaney C. O’Connell, Bruce J. Turner, Christopher H. Martin* 10 11 *Correspondence to: [email protected] 12 13 14 15 This PDF file includes: 16 17 Supplementary text 18 Figures S1 to S17 19 Tables S1 to S19 20 SI References 21 22 Other supplementary materials for this manuscript include the following: 23 24 Datasets S1 to S9 25 26 27 28 29 30 31 32 33 34 35 36 37 1 38 Table of Contents 1. Supplemental Materials and Methods Page 1.1 Sampling 4 1.2 Genomic Library Prep 5 1.3 De novo genome assembly and annotation 5 1.4 Population genotyping. 6 1.5 Population genetic analyses 8 1.6 Mutation rate estimation. 10 1.7 Demographic Inferences 12 1.8 Introgression in SSI specialists 13 1.9 Search for candidate adaptive alleles in SSI specialists 15 1.10 Introgression in outgroup generalist populations 18 1.11. Characterization of adaptive alleles through GO analysis 19 1.12 Characterization of adaptive alleles through genome-wide association mapping 19 1.13 Characterization of adaptive alleles through differential gene expression and QTL 22 analysis from previous studies 1.14 Timing of divergence among adaptive alleles 23 1.15 Timing of selective sweeps on adaptive alleles 26 2. Supplementary Results and Discussion 31 2 2.1 Spatiotemporal stages of adaption based on timing of divergence among adaptive alleles 31 2.2 Spatiotemporal stages of adaptation based on timing of selection on adaptive alleles 35 3.