Resolving the Ray-Finned Fish Tree of Life COMMENTARY Michael E
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Reef Fishes of the Bird's Head Peninsula, West
Check List 5(3): 587–628, 2009. ISSN: 1809-127X LISTS OF SPECIES Reef fishes of the Bird’s Head Peninsula, West Papua, Indonesia Gerald R. Allen 1 Mark V. Erdmann 2 1 Department of Aquatic Zoology, Western Australian Museum. Locked Bag 49, Welshpool DC, Perth, Western Australia 6986. E-mail: [email protected] 2 Conservation International Indonesia Marine Program. Jl. Dr. Muwardi No. 17, Renon, Denpasar 80235 Indonesia. Abstract A checklist of shallow (to 60 m depth) reef fishes is provided for the Bird’s Head Peninsula region of West Papua, Indonesia. The area, which occupies the extreme western end of New Guinea, contains the world’s most diverse assemblage of coral reef fishes. The current checklist, which includes both historical records and recent survey results, includes 1,511 species in 451 genera and 111 families. Respective species totals for the three main coral reef areas – Raja Ampat Islands, Fakfak-Kaimana coast, and Cenderawasih Bay – are 1320, 995, and 877. In addition to its extraordinary species diversity, the region exhibits a remarkable level of endemism considering its relatively small area. A total of 26 species in 14 families are currently considered to be confined to the region. Introduction and finally a complex geologic past highlighted The region consisting of eastern Indonesia, East by shifting island arcs, oceanic plate collisions, Timor, Sabah, Philippines, Papua New Guinea, and widely fluctuating sea levels (Polhemus and the Solomon Islands is the global centre of 2007). reef fish diversity (Allen 2008). Approximately 2,460 species or 60 percent of the entire reef fish The Bird’s Head Peninsula and surrounding fauna of the Indo-West Pacific inhabits this waters has attracted the attention of naturalists and region, which is commonly referred to as the scientists ever since it was first visited by Coral Triangle (CT). -

Shortest Recorded Vertebrate Lifespan Found in a Coral Reef Fish
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector Current Biology Vol 15 No 8 R288 monkeyflowers and other taxa are just 35 days, of which at least 10 helping to overcome this gap. Correspondence are taken to reach sexual maturity The ‘bottom up’ or genetic (Figure 1B). This provides the approach to studying speciation species with a remarkable three- has hunted down genes Shortest recorded week window in which to responsible for premating and reproduce and contribute to the postmating isolation, and then vertebrate next generation. shown that the gene sequences lifespan found in a Already constrained by time, the exhibit signatures of recent lifetime fecundity of E. sigillata is selection. But this approach has coral reef fish further restricted by small adult told us little about the nature of body sizes of 11–20 mm, limiting that selection. Is selection the number of eggs a female can divergent or has divergence Martial Depczynski and produce. Yet pygmy gobies are an occurred under uniform selection? David R. Bellwood incredibly successful group, Was selection in response to numbering some 70 species with environmental differences? Was it Extreme short lifespans are of a geographic distribution natural or sexual selection? interest because they mark encompassing reefs across the Finally, we still know little about current evolutionary boundaries Indian and Pacific Oceans [1]. To how mate preferences evolve and biological limits within which investigate lifetime fecundity, we within and between populations life’s essential tasks must be bred pygmy gobies in captivity. during the process of speciation. -

Taxonomic Research of the Gobioid Fishes (Perciformes: Gobioidei) in China
KOREAN JOURNAL OF ICHTHYOLOGY, Vol. 21 Supplement, 63-72, July 2009 Received : April 17, 2009 ISSN: 1225-8598 Revised : June 15, 2009 Accepted : July 13, 2009 Taxonomic Research of the Gobioid Fishes (Perciformes: Gobioidei) in China By Han-Lin Wu, Jun-Sheng Zhong1,* and I-Shiung Chen2 Ichthyological Laboratory, Shanghai Ocean University, 999 Hucheng Ring Rd., 201306 Shanghai, China 1Ichthyological Laboratory, Shanghai Ocean University, 999 Hucheng Ring Rd., 201306 Shanghai, China 2Institute of Marine Biology, National Taiwan Ocean University, Keelung 202, Taiwan ABSTRACT The taxonomic research based on extensive investigations and specimen collections throughout all varieties of freshwater and marine habitats of Chinese waters, including mainland China, Hong Kong and Taiwan, which involved accounting the vast number of collected specimens, data and literature (both within and outside China) were carried out over the last 40 years. There are totally 361 recorded species of gobioid fishes belonging to 113 genera, 5 subfamilies, and 9 families. This gobioid fauna of China comprises 16.2% of 2211 known living gobioid species of the world. This report repre- sents a summary of previous researches on the suborder Gobioidei. A recently diagnosed subfamily, Polyspondylogobiinae, were assigned from the type genus and type species: Polyspondylogobius sinen- sis Kimura & Wu, 1994 which collected around the Pearl River Delta with high extremity of vertebral count up to 52-54. The undated comprehensive checklist of gobioid fishes in China will be provided in this paper. Key words : Gobioid fish, fish taxonomy, species checklist, China, Hong Kong, Taiwan INTRODUCTION benthic perciforms: gobioid fishes to evolve and active- ly radiate. The fishes of suborder Gobioidei belong to the largest The gobioid fishes in China have long received little group of those in present living Perciformes. -

Patterns of Evolution in Gobies (Teleostei: Gobiidae): a Multi-Scale Phylogenetic Investigation
PATTERNS OF EVOLUTION IN GOBIES (TELEOSTEI: GOBIIDAE): A MULTI-SCALE PHYLOGENETIC INVESTIGATION A Dissertation by LUKE MICHAEL TORNABENE BS, Hofstra University, 2007 MS, Texas A&M University-Corpus Christi, 2010 Submitted in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY in MARINE BIOLOGY Texas A&M University-Corpus Christi Corpus Christi, Texas December 2014 © Luke Michael Tornabene All Rights Reserved December 2014 PATTERNS OF EVOLUTION IN GOBIES (TELEOSTEI: GOBIIDAE): A MULTI-SCALE PHYLOGENETIC INVESTIGATION A Dissertation by LUKE MICHAEL TORNABENE This dissertation meets the standards for scope and quality of Texas A&M University-Corpus Christi and is hereby approved. Frank L. Pezold, PhD Chris Bird, PhD Chair Committee Member Kevin W. Conway, PhD James D. Hogan, PhD Committee Member Committee Member Lea-Der Chen, PhD Graduate Faculty Representative December 2014 ABSTRACT The family of fishes commonly known as gobies (Teleostei: Gobiidae) is one of the most diverse lineages of vertebrates in the world. With more than 1700 species of gobies spread among more than 200 genera, gobies are the most species-rich family of marine fishes. Gobies can be found in nearly every aquatic habitat on earth, and are often the most diverse and numerically abundant fishes in tropical and subtropical habitats, especially coral reefs. Their remarkable taxonomic, morphological and ecological diversity make them an ideal model group for studying the processes driving taxonomic and phenotypic diversification in aquatic vertebrates. Unfortunately the phylogenetic relationships of many groups of gobies are poorly resolved, obscuring our understanding of the evolution of their ecological diversity. This dissertation is a multi-scale phylogenetic study that aims to clarify phylogenetic relationships across the Gobiidae and demonstrate the utility of this family for studies of macroevolution and speciation at multiple evolutionary timescales. -

Insights from Coral Reef Fishes
Ecology, 87(12), 2006, pp. 3119–3127 Ó 2006 by the Ecological Society of America EXTREMES, PLASTICITY, AND INVARIANCE IN VERTEBRATE LIFE HISTORY TRAITS: INSIGHTS FROM CORAL REEF FISHES 1 MARTIAL DEPCZYNSKI AND DAVID R. BELLWOOD ARC Centre of Excellence for Coral Reef Studies, School of Marine Biology, James Cook University, Townsville 4811 Australia Abstract. Life history theory predicts a range of directional generic responses in life history traits with increasing organism size. Among these are the relationships between size and longevity, mortality, growth rate, timing of maturity, and lifetime reproductive output. Spanning three orders of magnitude in size, coral reef fishes provide an ecologically diverse and species-rich vertebrate assemblage in which to test these generic responses. Here we examined these relationships by quantifying the life cycles of three miniature species of coral reef fish from the genus Eviota (Gobiidae) and compared their life history characteristics with other reef fish species. We found that all three species of Eviota have life spans of ,100 days, suffer high daily mortality rates of 7–8%, exhibit rapid linear growth, and matured at an earlier than expected size. Although lifetime reproductive output was low, consistent with their small body sizes, short generation times of 47–74 days help overcome low individual fecundity and appear to be a critical feature in maintaining Eviota populations. Comparisons with other coral reef fish species showed that Eviota species live on the evolutionary margins of life history possibilities for vertebrate animals. This addition of demographic information on these smallest size classes of coral reef fishes greatly extends our knowledge to encompass the full size spectrum and highlights the potential for coral reef fishes to contribute to vertebrate life history studies. -
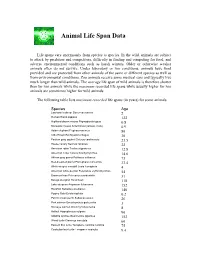
Animal Life Span Data
Animal Life Span Data Life spans vary enormously from species to species. In the wild, animals are subject to attack by predators and competitors, difficulty in finding and competing for food, and adverse environmental conditions such as harsh winters. Older or otherwise weaker animals often do not survive. Under laboratory or zoo conditions, animals have food provided and are protected from other animals of the same or different species as well as from environmental conditions. Zoo animals receive some medical care and typically live much longer than wild animals. The average life span of wild animals is therefore shorter than for zoo animals while the maximum recorded life spans while usually higher for zoo animals are sometimes higher for wild animals. The following table lists maximum recorded life spans (in years) for some animals. Species Age Laxmann’s shrew Sorex caecutiens 2 Human Homo sapiens 122 Highland desert mouse Eligmodontia typus 0.8 Marsupial mouse Antechinus (various, male) 0.9 Asian elephant Elephas maximus 80 Little Brown Bat Myotis lucifugus 30 Eastern gray squirrel Sciurus carolinensis 23.5 House canary Serinus canarius 22 American robin Turdus migratorius 12.8 American Crow Corvus brachyrhynchos 14.6 African gray parrot Psittacus erithacus 73 Red-breasted parrot Poicephalus rufiventris 33.4 White-winged crossbill Loxia leucoptera 4 American white pelican Pelecanus erythrorhynchos 54 Brown pelican Pelecanus occidentalis 31 Beluga sturgeon Huso huso 118 Lake sturgeon Acipenser fulvescens 152 Rockfish Sebastes aleutianus 140 Pygmy Gobi Eviota sigillata 0.2 Pacific ocean perch Sebastes alutus 26 Pink salmon Oncorhynchus gorbuscha 3 Sockeye salmon Oncorhynchus nerka 8 Halibut Hippoglossus vulgaris 90 Aldabra tortoise Geochelone gigantea 152 Wood turtle Clemmys insculpta 60 Eastern box turtle Terrapene carolina carolina 75 Coahuilan box turtle Terrapene coahuila 9.4 One point of this table was to illustrate that very similar species sometimes have dramatically different life spans. -

Short‐Lived Fishes: Annual and Multivoltine Strategies
Received: 12 October 2020 | Revised: 10 December 2020 | Accepted: 10 December 2020 DOI: 10.1111/faf.12535 ORIGINAL ARTICLE Short- lived fishes: Annual and multivoltine strategies Jakub Žák1,2 | Milan Vrtílek1 | Matej Polačik1 | Radim Blažek1,3 | Martin Reichard1,3,4 1Institute of Vertebrate Biology of the Czech Academy of Sciences, Brno, Czech Republic Abstract 2Department of Zoology, Faculty of Science, The diversity of life histories across the animal kingdom is enormous, with direct Charles University, Prague, Czech Republic consequences for the evolution of lifespans. Very short lifespans (maximum shorter 3Department of Botany and Zoology, Faculty of Science, Masaryk University, Brno, Czech than 1 year in their natural environment) have evolved in several vertebrate lineages. Republic We review short- lived fish species which complete either single (annual/univoltine) 4 Department of Ecology and Vertebrate or multiple (multivoltine) generations within a year. We summarize the commonali- Zoology, University of Łódź, Łódź, Poland ties and particulars of their biology. Apart from annual killifishes (with >350 spe- Correspondence cies), we detected 60 species with validated lifespan shorter than 1 year in their Martin Reichard, Institute of Vertebrate Biology of the Czech Academy of Sciences, natural environment. Considering the low number of reports on fish lifespan (<5% Květná 8, 603 00, Brno, Czech Republic. of 30,000+ fish species; 1,543 species), the total number of short- lived fish species Email: [email protected] may be relatively high (>1,200 species). Short- lived fish species are scattered across Funding information 12 orders, indicating that short lifespan is not a phylogenetically conserved trait but Czech Science Foundation, Grant/Award Number: 19- 01781S; Department of rather evolves under specific ecological conditions. -

The Productivity of Coral Reef Fishes
ResearchOnline@JCU This file is part of the following work: Morais Araujo, Renato (2020) The productivity of coral reef fishes. PhD Thesis, James Cook University. Access to this file is available from: https://doi.org/10.25903/c2h5%2Df063 Copyright © 2020 Renato Morais Araujo. The author has certified to JCU that they have made a reasonable effort to gain permission and acknowledge the owners of any third party copyright material included in this document. If you believe that this is not the case, please email [email protected] The productivity of coral reef fishes Renato Morais Araujo February 2020 For the Degree of Doctor of Philosophy College of Science and Engineering ARC Centre of Excellence for Coral Reef Studies James Cook University Dedico esta tese ao Vovô Manoel, avô, pai, herói ii Acknowledgements I thank my supervisor, David Bellwood, for continued guidance, counselling, and unconditional availability and support throughout the thesis; for those long hours of brain-draining, incredibly insightful meetings, and for not minding my artful representations of him in presentations. Thanks also to Sean Connolly for his disproportional contribution to the theoretical development of this thesis, which happened in the context of a particularly challenging chapter. Thanks to all who kindly helped me during field work: Pauline Narvaez, Victor Huertas, Ale Siqueira, Chris Hemingson, Sterling Tebbett, Rob Streit, Joshua Phua, Sam Shu Quin and Sabrina Inderbitzi. Special thanks to ‘my’ rusty chain field crew, Pauline, Victor and Ale, for taking over big responsibilities while I eternally counted 68,656 big or small fish. You tamed Quaddie and marinated chains (just enough rustiness), spending days at depths deeper than the height of a (short) person. -

Tonga SUMA Report
BIOPHYSICALLY SPECIAL, UNIQUE MARINE AREAS OF TONGA EFFECTIVE MANAGEMENT Marine and coastal ecosystems of the Pacific Ocean provide benefits for all people in and beyond the region. To better understand and improve the effective management of these values on the ground, Pacific Island Countries are increasingly building institutional and personal capacities for Blue Planning. But there is no need to reinvent the wheel, when learning from experiences of centuries of traditional management in Pacific Island Countries. Coupled with scientific approaches these experiences can strengthen effective management of the region’s rich natural capital, if lessons learnt are shared. The MACBIO project collaborates with national and regional stakeholders towards documenting effective approaches to sustainable marine resource management and conservation. The project encourages and supports stakeholders to share tried and tested concepts and instruments more widely throughout partner countries and the Oceania region. This report outlines the process undertaken to define and describe the special, unique marine areas of Tonga. These special, unique marine areas provide an important input to decisions about, for example, permits, licences, EIAs and where to place different types of marine protected areas, locally managed marine areas and Community Conservation Areas in Tonga. For a copy of all reports and communication material please visit www.macbio-pacific.info. MARINE ECOSYSTEM MARINE SPATIAL PLANNING EFFECTIVE MANAGEMENT SERVICE VALUATION BIOPHYSICALLY SPECIAL, UNIQUE MARINE AREAS OF TONGA AUTHORS: Ceccarelli DM1, Wendt H2, Matoto AL3, Fonua E3, Fernandes L2 SUGGESTED CITATION: Ceccarelli DM, Wendt H, Matoto AL, Fonua E and Fernandes L (2017) Biophysically special, unique marine areas of Tonga. MACBIO (GIZ, IUCN, SPREP), Suva. -

NBSREA Design Cvrs V2.Pub
February 2009 TNC Pacific Island Countries Report No 1/09 Rapid Ecological Assessment Northern Bismarck Sea Papua New Guinea Technical report of survey conducted August 13 to September 7, 2006 Edited by: Richard Hamilton, Alison Green and Jeanine Almany Supported by: AP Anonymous February 2009 TNC Pacific Island Countries Report No 1/09 Rapid Ecological Assessment Northern Bismarck Sea Papua New Guinea Technical report of survey conducted August 13 to September 7, 2006 Edited by: Richard Hamilton, Alison Green and Jeanine Almany Published by: The Nature Conservancy, Indo-Pacific Resource Centre Author Contact Details: Dr. Richard Hamilton, 51 Edmondstone Street, South Brisbane, QLD 4101 Australia Email: [email protected] Suggested Citation: Hamilton, R., A. Green and J. Almany (eds.) 2009. Rapid Ecological Assessment: Northern Bismarck Sea, Papua New Guinea. Technical report of survey conducted August 13 to September 7, 2006. TNC Pacific Island Countries Report No. 1/09. © 2009, The Nature Conservancy All Rights Reserved. Reproduction for any purpose is prohibited without prior permission. Cover Photo: Manus © Gerald Allen ISBN 9980-9964-9-8 Available from: Indo-Pacific Resource Centre The Nature Conservancy 51 Edmondstone Street South Brisbane, QLD 4101 Australia Or via the worldwide web at: conserveonline.org/workspaces/pacific.island.countries.publications ii Foreword Manus and New Ireland provinces lie north of the Papua New Guinea mainland in the Bismarck Archipelago. More than half of the local communities in our provinces are coastal inhabitants, who for thousands of years have depended on marine resources for their livelihood. For coastal communities survival and prosperity is integrally linked to healthy marine ecosystems. -

Resolving the Ray-Finned Fish Tree of Life COMMENTARY Michael E
COMMENTARY Resolving the ray-finned fish tree of life COMMENTARY Michael E. Alfaroa,1 The Challenge of Reconstructing Fish Phylogeny responsible for generating the trees in the first place When it comes to vertebrate evolutionary history, has emerged. In PNAS, Hughes et al. (12), use over our understanding of lobe-finned fishes—the branch 1,000 new markers using phylogenomic and tran- of the vertebrate tree leading to coelacanths plus scriptomic techniques to reconstruct the evolutionary the tetrapods (amphibians, turtles, birds, crocodiles, history of ray-finned fishes. Hughes et al.’s phylogeny lizards, snakes, and mammals)—far outstrips our reveals many key features of the backbone of the ray- knowledge of ray-finned fishes (Actinopterygii). Acti- finned fish tree of life and provides the most com- nopterygians exhibit extraordinary species richness prehensive evolutionary framework to date for this (>33,000 described species) and have evolved a stag- major radiation of vertebrates. gering diversity in morphology and ecology over their Phylogenomics has revolutionized evolutionary 400+ million y history. Ray-finned fishes include some reconstruction by vastly expanding both the number of the smallest vertebrates [the adult cyprinid Paedo- of loci that can be interrogated across nonmodel or- cypris progenetica measure just under 8 mm (1)], some ganisms, as well as the efficiency with which the data of the largest (adult ocean sunfish weigh more than may be collected (13). The most recent multilocus 2,000 kg), some of the longest (oarfishes may reach a studies that underlie our current understanding of the length of more than 13 m), some of the longest lived ray-finned fish tree of life are based upon 10– [rougheye rockfishes, Sebastes aleutianus, may live for 20 markers (e.g., refs. -

Life Strategies in the Long-Lived Bivalve Arctica Islandica on a Latitudinal Climate Gradient – Environmental Constraints and Evolutionary Adaptations
Life strategies in the long-lived bivalve Arctica islandica on a latitudinal climate gradient – Environmental constraints and evolutionary adaptations Lebensstrategien der langlebigen Muschel Arctica islandica, untersucht an Populationen entlang eines Klimagradienten – Umwelteinflüsse und evolutionäre Anpassungen Dissertation zur Erlangung des Grades eines Doktors der Naturwissenschaften -Dr. rer. nat.- Fachbereich 2 Biologie/Chemie Universität Bremen vorgelegt von Julia Strahl Bremen März 2011 Prüfungsausschuss: 1. Gutachter: Prof. Dr. Ralf Dringen (Zentrum für Biomolekulare Interaktionen Bremen, Universität Bremen) 2. Gutachter: PD Dr. Doris Abele (Funktionelle Ökologie, Alfred-Wegener-Institut für Polar- und Meeresforschung, Bremerhaven) 1. Prüfer: Prof. Dr. Thomas Brey (Funktionelle Ökologie, Alfred-Wegener-Institut für Polar- und Meeresforschung, Bremerhaven) 2. Prüfer: Prof. Dr. Kai Bischof (Marine Botanik, Universität Bremen) CONTENTS FREQUENTLY USED ABBREVATIONS I SUMMARY III ZUSAMMENFASSUNG V 1. INTRODUCTION 1 1.1. Bivalves as models in aging research 1 1.2. Why is Arctica islandica an interesting model in aging research? 2 1.3. What is aging? 4 1.4. Physiological parameters involved in the aging process 5 1.5. Is the aging process in ectotherms related to reactive oxygen species formation? 6 1.6. Cellular maintenance and longevity 7 1.7. Metabolic rate depression and longevity 9 1.8. Possible role of nitric oxide in metabolic rate depression 10 1.9. Metabolic rate depression, anaerobiosis and recovery from anoxia 10 1.10. Aims of the thesis 13 2. MATERIALS AND METHODS – A GENERAL OVERVIEW 14 2.1. Investigated species and sampling locations 14 2.2. Experimental studies 15 2.2.1. Incubation experiment for the determination of cell-turnover rates 15 2.2.2.