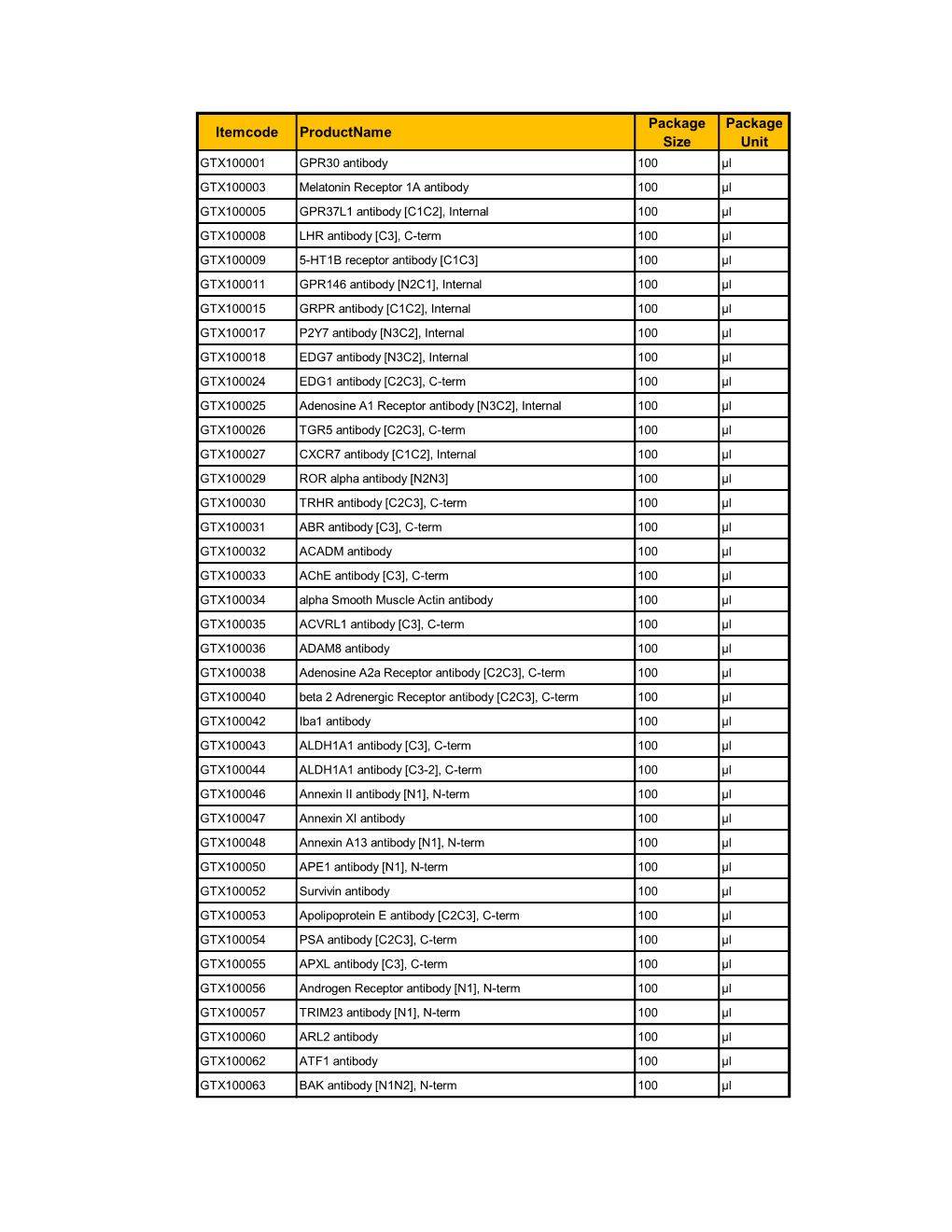
Itemcode Productname Package Size Package Unit
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Applying Screening Techniques to Two Orphan Gpcrs
Universidade de Lisboa Faculdade de Farmácia Deorphanization of receptors: Applying screening techniques to two orphan GPCRs Ana Catarina Rufas da Silva Santos Mestrado Integrado em Ciências Farmacêuticas 2019 Universidade de Lisboa Faculdade de Farmácia Deorphanization of receptors: Applying screening techniques to two orphan GPCRs Ana Catarina Rufas da Silva Santos Monografia de Mestrado Integrado em Ciências Farmacêuticas apresentada à Universidade de Lisboa através da Faculdade de Farmácia Orientadora: Ghazl Al Hamwi, PhD Student Co-Orientadora: Professora Doutora Elsa Maria Ribeiro dos Santos Anes, Professora Associada com Agregação em Microbiologia 2019 Abstract G-Protein Coupled Receptors represent one of the largest families of cellular receptors discovered and one of the main sources of attractive drug targets. In contrast, it also has a large number of understudied or orphan receptors. Pharmacological assays such as β-Arrestin recruitment assays, are one of the possible approaches for deorphanization of receptors. In this work, I applied the assay system previously mentioned to screen compounds in two orphan receptors, GRP37 and MRGPRX3. GPR37 has been primarily associated with a form of early onset Parkinsonism due to its’ expression patterns, and physiological role as substrate to ubiquitin E3, parkin. Although extensive literature regarding this receptor is available, the identification of a universally recognized ligand has not yet been possible. Two compounds were proposed as ligands, but both were met with controversy. These receptor association with Autosomal Recessive Juvenile Parkinson positions it as a very attractive drug target, and as such its’ deorphanization is a prime objective for investigators in this area. Regarding MRGPRX3 information is much scarcer. -

Supporting Information
Supporting Information Koyanagi et al. 10.1073/pnas.0806215105 SI Materials and Methods CNGB, NM137763; human CNGB1, NM001297; human Amino Acid Sequences. The accession numbers of amino acid CNGB3, NM 019098; box jellyfish CNG, AB435552; fruit fly sequences used for analyses are as follows: Box jellyfish opsin, CNG4, NM 167441; fruit fly CNG3, NM 137871; human AB435549; sea anemone opsin, BR000662; hydra opsin, Con- CNGA4, NM 001037329; fruit fly CNGA, NM 057768; human CNGA1, NM000087; human CNGA2, NM005140; human tig39347:487820–488855 (http://hydrazome.metazome.net); hy- drozaon jellyfish opsin, AB332435; human encephalopsin, CNGA3, NM 001298. AF140242; ragworm c-opsin, AY692353; human rhodopsin, Western Blot Analysis. The proteins extracted from half a rhopalia U49742; human blue, M13299; human red, Z68193; lamprey were separated by 12% SDS/PAGE, transferred onto a PVDF parapinopsin, AB116380; lizard parietopsin, DQ100320; am- membrane, and incubated with 1:500 diluted anti-box jellyfish phioxus peropsin, AB050610; human peropsin, AF012270; hu- opsin antiserum. Visualization was carried out by the ABC man rgr, U15790; squid retinochrome, X57143; human melan- method (Vectastain) and by staining with 3,3Ј-diaminobenzidine opsin, AF147788; amphioxus melanopsin, AB205400; squid (Sigma). rhodopsin, X70498; fruit fly Rh1, K02315; human neuropsin, AY377391; amphioxus rhodopsin, AB050606; scallop Go- In Situ Hybridization. Digoxigenin-labeled antisense and sense RNA rhodopsin, AB006455; human muscarinic acetylcholine receptor probes for the box jellyfish opsin were synthesized by using the DIG M1 (CHRM1), NM000738; human melatonin receptor 1A RNA labeling kit (Roche). Sections were pretreated with protein- (MTNR1A), NM005958; Arabidopsis AKT1, U06745; Arabi- ase K and hybridized with each RNA probe. -

Cytoplasmic Activation-Induced Cytidine Deaminase (AID) Exists in Stoichiometric Complex with Translation Elongation Factor 1Α (Eef1a)
Cytoplasmic activation-induced cytidine deaminase (AID) exists in stoichiometric complex with translation elongation factor 1α (eEF1A) Julien Häsler, Cristina Rada, and Michael S. Neuberger1 Medical Research Council Laboratory of Molecular Biology, Cambridge CB2 0QH, United Kingdom Edited by Frederick W. Alt, Howard Hughes Medical Institute, Harvard Medical School, Children’s Hospital Immune Disease Institute, Boston, MA, and approved October 12, 2011 (received for review April 27, 2011) Activation-induced cytidine deaminase (AID) is a B lymphocyte- results reveal that endogenous cytoplasmic AID partakes in a specific DNA deaminase that acts on the Ig loci to trigger antibody complex containing stoichiometric quantities of translation elon- gene diversification. Most AID, however, is retained in the cyto- gation factor 1α (eEF1A), with this association likely implicated in plasm and its nuclear abundance is carefully regulated because the regulation of AID’s intracellular trafficking. off-target action of AID leads to cancer. The nature of the cytosolic AID complex and the mechanisms regulating its release from the Results cytoplasm and import into the nucleus remain unknown. Here, we Flag-Tagging the Endogenous AID Locus in DT40 Cells. We generated show that cytosolic AID in DT40 B cells is part of an 11S complex derivatives of the DT40 B-cell line in which the endogenous AID and, using an endogenously tagged AID protein to avoid overex- locus was modified so as to incorporate a single Flag tag at the pression artifacts, that it is bound in good stoichiometry to the AID N terminus. To allow targeting of both alleles, one targeting translation elongation factor 1 alpha (eEF1A). -

Mast Cells Promote Seasonal White Adipose Beiging in Humans
Diabetes Volume 66, May 2017 1237 Mast Cells Promote Seasonal White Adipose Beiging in Humans Brian S. Finlin,1 Beibei Zhu,1 Amy L. Confides,2 Philip M. Westgate,3 Brianna D. Harfmann,1 Esther E. Dupont-Versteegden,2 and Philip A. Kern1 Diabetes 2017;66:1237–1246 | DOI: 10.2337/db16-1057 Human subcutaneous (SC) white adipose tissue (WAT) localized to the neck and thorax of humans (4–8), and in a increases the expression of beige adipocyte genes in the process known as beiging (9), UCP1-positive adipocytes winter. Studies in rodents suggest that a number of form in subcutaneous (SC) white adipose tissue (WAT) immune mediators are important in the beiging response. (10). Beige adipocytes have unique developmental origins, We studied the seasonal beiging response in SC WAT gene signatures, and functional properties, including being from lean humans. We measured the gene expression of highly inducible to increase UCP1 in response to catechol- various immune cell markers and performed multivariate amines (9,11–13). Although questions exist about whether analysis of the gene expression data to identify genes beige fat can make a meaningful contribution to energy OBESITY STUDIES that predict UCP1. Interleukin (IL)-4 and, unexpectedly, expenditure in humans (reviewed in Porter et al. [14]), the mast cell marker CPA3 predicted UCP1 gene expres- the induction of beige fat in rodent models is associated sion. Therefore, we investigated the effects of mast with increased energy expenditure and improved glucose cells on UCP1 induction by adipocytes. TIB64 mast cells homeostasis (13). responded to cold by releasing histamine and IL-4, and this medium stimulated UCP1 expression and lipolysis by Activation of the sympathetic nervous system by cold 3T3-L1 adipocytes. -

Evolution, Expression and Meiotic Behavior of Genes Involved in Chromosome Segregation of Monotremes
G C A T T A C G G C A T genes Article Evolution, Expression and Meiotic Behavior of Genes Involved in Chromosome Segregation of Monotremes Filip Pajpach , Linda Shearwin-Whyatt and Frank Grützner * School of Biological Sciences, The University of Adelaide, Adelaide, SA 5005, Australia; fi[email protected] (F.P.); [email protected] (L.S.-W.) * Correspondence: [email protected] Abstract: Chromosome segregation at mitosis and meiosis is a highly dynamic and tightly regulated process that involves a large number of components. Due to the fundamental nature of chromosome segregation, many genes involved in this process are evolutionarily highly conserved, but duplica- tions and functional diversification has occurred in various lineages. In order to better understand the evolution of genes involved in chromosome segregation in mammals, we analyzed some of the key components in the basal mammalian lineage of egg-laying mammals. The chromosome passenger complex is a multiprotein complex central to chromosome segregation during both mitosis and meio- sis. It consists of survivin, borealin, inner centromere protein, and Aurora kinase B or C. We confirm the absence of Aurora kinase C in marsupials and show its absence in both platypus and echidna, which supports the current model of the evolution of Aurora kinases. High expression of AURKBC, an ancestor of AURKB and AURKC present in monotremes, suggests that this gene is performing all necessary meiotic functions in monotremes. Other genes of the chromosome passenger complex complex are present and conserved in monotremes, suggesting that their function has been preserved Citation: Pajpach, F.; in mammals. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -

Expression of Melatonin Receptors 1A/1B, Calmodulin and Estrogen Receptor 2 in Deep Paravertebral Muscles Revisited
MOLECULAR MEDICINE REPORTS 14: 5719-5724, 2016 Etiopathogenesis of adolescent idiopathic scoliosis: Expression of melatonin receptors 1A/1B, calmodulin and estrogen receptor 2 in deep paravertebral muscles revisited JOSEF ZAMECNIK1*, LENKA KRSKOVA1*, JAROMIR HACEK1, IVANA STETKAROVA2 and MARTIN KRBEC3 1Department of Pathology and Molecular Medicine, 2nd Faculty of Medicine, Charles University in Prague and University Hospital Motol, 15006 Prague; 2Department of Neurology, Charles University; 3Department of Orthopedics and Traumatology, 3rd Faculty of Medicine, Charles University in Prague and University Hospital Královské Vinohrady, 10034 Prague, Czech Republic Received October 28, 2015; Accepted October 11, 2016 DOI: 10.3892/mmr.2016.5927 Abstract. The pathogenesis of adolescent idiopathic scoliosis addition, no difference in expression was detected between the (AIS), including the associated local changes in deep para- patients with AIS and the controls. With regards to MTNR1A vertebral muscles, is poorly understood. The asymmetric and MTNR1B, their expression was very weak in paravertebral expression of several molecules involved in the melatonin muscles, and in the majority of cases their expression could signaling pathway, including melatonin receptors 1A/1B not be detected by repeated RT-qPCR analysis. Therefore, (MTNR1A/MTNR1B), estrogen receptor 2 (ESR2) and these data do not support the previously suggested role of the calmodulin (CALM1), has previously been suggested to be asymmetric expression of molecules involved in the melatonin associated with AIS. However, this hypothesis is based on signaling pathway in deep paravertebral muscles in the patho- single studies in which the data were obtained by different genesis of AIS. methodological approaches. Therefore, to evaluate the symmetry of the mRNA expression levels of these molecules, Introduction 18 patients with AIS and 10 non-scoliotic controls were enrolled in the present study. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Investigation of Candidate Genes and Mechanisms Underlying Obesity
Prashanth et al. BMC Endocrine Disorders (2021) 21:80 https://doi.org/10.1186/s12902-021-00718-5 RESEARCH ARTICLE Open Access Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules G. Prashanth1 , Basavaraj Vastrad2 , Anandkumar Tengli3 , Chanabasayya Vastrad4* and Iranna Kotturshetti5 Abstract Background: Obesity associated type 2 diabetes mellitus is a metabolic disorder ; however, the etiology of obesity associated type 2 diabetes mellitus remains largely unknown. There is an urgent need to further broaden the understanding of the molecular mechanism associated in obesity associated type 2 diabetes mellitus. Methods: To screen the differentially expressed genes (DEGs) that might play essential roles in obesity associated type 2 diabetes mellitus, the publicly available expression profiling by high throughput sequencing data (GSE143319) was downloaded and screened for DEGs. Then, Gene Ontology (GO) and REACTOME pathway enrichment analysis were performed. The protein - protein interaction network, miRNA - target genes regulatory network and TF-target gene regulatory network were constructed and analyzed for identification of hub and target genes. The hub genes were validated by receiver operating characteristic (ROC) curve analysis and RT- PCR analysis. Finally, a molecular docking study was performed on over expressed proteins to predict the target small drug molecules. Results: A total of 820 DEGs were identified between -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Functional Genomics Atlas of Synovial Fibroblasts Defining Rheumatoid Arthritis
medRxiv preprint doi: https://doi.org/10.1101/2020.12.16.20248230; this version posted December 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Functional genomics atlas of synovial fibroblasts defining rheumatoid arthritis heritability Xiangyu Ge1*, Mojca Frank-Bertoncelj2*, Kerstin Klein2, Amanda Mcgovern1, Tadeja Kuret2,3, Miranda Houtman2, Blaž Burja2,3, Raphael Micheroli2, Miriam Marks4, Andrew Filer5,6, Christopher D. Buckley5,6,7, Gisela Orozco1, Oliver Distler2, Andrew P Morris1, Paul Martin1, Stephen Eyre1* & Caroline Ospelt2*,# 1Versus Arthritis Centre for Genetics and Genomics, School of Biological Sciences, Faculty of Biology, Medicine and Health, The University of Manchester, Manchester, UK 2Department of Rheumatology, Center of Experimental Rheumatology, University Hospital Zurich, University of Zurich, Zurich, Switzerland 3Department of Rheumatology, University Medical Centre, Ljubljana, Slovenia 4Schulthess Klinik, Zurich, Switzerland 5Institute of Inflammation and Ageing, University of Birmingham, Birmingham, UK 6NIHR Birmingham Biomedical Research Centre, University Hospitals Birmingham NHS Foundation Trust, University of Birmingham, Birmingham, UK 7Kennedy Institute of Rheumatology, University of Oxford Roosevelt Drive Headington Oxford UK *These authors contributed equally #corresponding author: [email protected] NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. 1 medRxiv preprint doi: https://doi.org/10.1101/2020.12.16.20248230; this version posted December 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity.