Expression of Melatonin Receptors 1A/1B, Calmodulin and Estrogen Receptor 2 in Deep Paravertebral Muscles Revisited
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supporting Information
Supporting Information Koyanagi et al. 10.1073/pnas.0806215105 SI Materials and Methods CNGB, NM137763; human CNGB1, NM001297; human Amino Acid Sequences. The accession numbers of amino acid CNGB3, NM 019098; box jellyfish CNG, AB435552; fruit fly sequences used for analyses are as follows: Box jellyfish opsin, CNG4, NM 167441; fruit fly CNG3, NM 137871; human AB435549; sea anemone opsin, BR000662; hydra opsin, Con- CNGA4, NM 001037329; fruit fly CNGA, NM 057768; human CNGA1, NM000087; human CNGA2, NM005140; human tig39347:487820–488855 (http://hydrazome.metazome.net); hy- drozaon jellyfish opsin, AB332435; human encephalopsin, CNGA3, NM 001298. AF140242; ragworm c-opsin, AY692353; human rhodopsin, Western Blot Analysis. The proteins extracted from half a rhopalia U49742; human blue, M13299; human red, Z68193; lamprey were separated by 12% SDS/PAGE, transferred onto a PVDF parapinopsin, AB116380; lizard parietopsin, DQ100320; am- membrane, and incubated with 1:500 diluted anti-box jellyfish phioxus peropsin, AB050610; human peropsin, AF012270; hu- opsin antiserum. Visualization was carried out by the ABC man rgr, U15790; squid retinochrome, X57143; human melan- method (Vectastain) and by staining with 3,3Ј-diaminobenzidine opsin, AF147788; amphioxus melanopsin, AB205400; squid (Sigma). rhodopsin, X70498; fruit fly Rh1, K02315; human neuropsin, AY377391; amphioxus rhodopsin, AB050606; scallop Go- In Situ Hybridization. Digoxigenin-labeled antisense and sense RNA rhodopsin, AB006455; human muscarinic acetylcholine receptor probes for the box jellyfish opsin were synthesized by using the DIG M1 (CHRM1), NM000738; human melatonin receptor 1A RNA labeling kit (Roche). Sections were pretreated with protein- (MTNR1A), NM005958; Arabidopsis AKT1, U06745; Arabi- ase K and hybridized with each RNA probe. -

Quantigene Flowrna Probe Sets Currently Available
QuantiGene FlowRNA Probe Sets Currently Available Accession No. Species Symbol Gene Name Catalog No. NM_003452 Human ZNF189 zinc finger protein 189 VA1-10009 NM_000057 Human BLM Bloom syndrome VA1-10010 NM_005269 Human GLI glioma-associated oncogene homolog (zinc finger protein) VA1-10011 NM_002614 Human PDZK1 PDZ domain containing 1 VA1-10015 NM_003225 Human TFF1 Trefoil factor 1 (breast cancer, estrogen-inducible sequence expressed in) VA1-10016 NM_002276 Human KRT19 keratin 19 VA1-10022 NM_002659 Human PLAUR plasminogen activator, urokinase receptor VA1-10025 NM_017669 Human ERCC6L excision repair cross-complementing rodent repair deficiency, complementation group 6-like VA1-10029 NM_017699 Human SIDT1 SID1 transmembrane family, member 1 VA1-10032 NM_000077 Human CDKN2A cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) VA1-10040 NM_003150 Human STAT3 signal transducer and activator of transcripton 3 (acute-phase response factor) VA1-10046 NM_004707 Human ATG12 ATG12 autophagy related 12 homolog (S. cerevisiae) VA1-10047 NM_000737 Human CGB chorionic gonadotropin, beta polypeptide VA1-10048 NM_001017420 Human ESCO2 establishment of cohesion 1 homolog 2 (S. cerevisiae) VA1-10050 NM_197978 Human HEMGN hemogen VA1-10051 NM_001738 Human CA1 Carbonic anhydrase I VA1-10052 NM_000184 Human HBG2 Hemoglobin, gamma G VA1-10053 NM_005330 Human HBE1 Hemoglobin, epsilon 1 VA1-10054 NR_003367 Human PVT1 Pvt1 oncogene homolog (mouse) VA1-10061 NM_000454 Human SOD1 Superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) -

Genetic Variations of Melatonin Receptor Type 1A Are Associated with the Clinicopathologic Development of Urothelial Cell Carcin
Int. J. Med. Sci. 2017, Vol. 14 1130 Ivyspring International Publisher International Journal of Medical Sciences 2017; 14(11): 1130-1135. doi: 10.7150/ijms.20629 Research Paper Genetic Variations of Melatonin Receptor Type 1A are Associated with the Clinicopathologic Development of Urothelial Cell Carcinoma Yung-Wei Lin1, 2, Shian-Shiang Wang3, 4, 5, Yu-Ching Wen2, 6, Min-Che Tung1, 7, Liang-Ming Lee2, 6, Shun-Fa Yang5, 8, Ming-Hsien Chien1, 9 1. Graduate Institute of Clinical Medicine, Taipei Medical University, Taipei, Taiwan; 2. Department of Urology, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan; 3. Division of Urology, Department of Surgery, Taichung Veterans General Hospital, Taichung, Taiwan; 4. School of Medicine, Chung Shan Medical University, Taichung, Taiwan; 5. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan; 6. Department of Urology, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan; 7. Department of Surgery, Tungs' Taichung Metro Harbor Hospital, Taichung, Taiwan; 8. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan; 9. Department of Medical Education and Research, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan. Corresponding authors: Ming-Hsien Chien, PhD, Graduate Institute of Clinical Medicine, Taipei Medical University, 250 Wu-Hsing Street, Taipei 11031, Taiwan; Phone: 886-2-27361661, ext. 3237; Fax: 886-2-27390500; E-mail: [email protected] or Shun-Fa Yang, PhD, Institute of Medicine, Chung Shan Medical University, 110 Chien-Kuo N. Road, Section 1, Taichung 402, Taiwan; Phone: 886-4-2473959, ext. 34253; Fax: 886-4-24723229; E-mail: [email protected] © Ivyspring International Publisher. -

Neurotransmission Alphabetical 28/7/05 15:52 Page 1
neuroscience - Neurotransmission alphabetical 28/7/05 15:52 Page 1 Abcam’s range of Neuroscience receptor, channel and ligand antibodies includes over 320 tried and tested products www.abcam.com Neurotransmission - receptor and channel signaling www.abcam.com GPCRs TYROSINE KINASES ION CHANNELS Neurotrophins rin u BDNF rt NGF u NT-3 Glutamate GABA ACh GDNF Ne Artemin Persepin NT-4/5 Glutamate Anandamide ATP Glycine ␥ 2-AG 1 2 3 4 / ␣ ␣ ␣ ␣ R / u A GFR GFR GFR GFR NTR mGL X1, 2, 3 CB1/2 2 EphA/B* Ephrin RET TrkA/B/C p75 AMPA/KainateNMDAR P GABA GlyR nAChR IP3 released Lyn Ephexin Homer DAG Adenylate Fyn PI3K Gr Src PSD95 Rapsin i PD2RGS3 Shc Syntrophin b K PLC Gi/q cyclase Gi 4 NcK PI3 Shc + 2+ 2+ PI3K PLC ␥ Na Ca Ca P PI3K Grb2 PLC/␥ PKC nNOS ␥ FAK PiP3/4 SOS MEK CaM Cl- Cl- ER NFkB PDEI PKA Rac RhoA RasGAP Src PTEN PKC Ras GTPase ERK CaMK 2+ PKK2 Calcineurin Ca released ROCK ERK FRS2 AKT/PKB Raf JNK nNOS SynGAP Grb2 actin MEK 1/2 P Ser133 Inhibition Cl- Src SOS Cell survival CREB of signaling RAS P CREB ERK 1/2 P P MAPK GSK3 ELK RSK NFB CREB CREB gene Long term Cytoskeleton transcription synaptic dynamics, neurite ASK1 Bad CRE plasticity (LTP) P P extension CREB CREB Short term MKK3/6 Apoptosis plasticity Others Other GPCRs - ARA9 ACh (Muscarinic): M1, M2, M3, M4 CRE p38MAPK c-fos Potassium - ASIC, ASIC3 Adrenalin: Alpha1b, 1c, 1d - KChIP2 - DDR 1, 2 ATP: P2Y MSK1 c-j c-fos gene - KCNQ 3, 5 - ENSA Dopamine: D1, D4 u transcription - Kv beta 2 - GJB1 n GABA: GABAB receptor - Kv 1.2 - SLC31A1 ␦ P P Opioid: µ, , CREB CREB AP1 - Kv -

Initial, Transient, and Specific Interaction Between G Protein
Sato T. et al. Medical Research Archives, vol. 6, issue 9, September 2018 Page 1 of 25 ARTICLE Initial, transient, and specific interaction between G protein-coupled receptor and target G protein in parallel signal processing: a case of olfactory discrimination of cancer-induced odors Takaaki Sato1, Mutsumi Matsukawa2, Yoichi Mizutani3, Toshio Iijima4, Hiroyoshi Matsumura5 Authors’ affiliations: 1 Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology, Osaka, Japan 2 Division of Anatomical Science, Department of Functional Morphology, Nihon University School of Medicine, Tokyo, Japan 3 Department of Medical Engineering, Faculty of Health Science, Aino University, Osaka, Japan 4 Graduate School of Life Sciences, Tohoku University, Sendai, Japan 5 College of Life Sciences, Ritsumeikan University, Kusatsu, Japan * Corresponding author: Takaaki Sato, Biomedical Research Institute, National Institute of Ad- vanced Industrial Science and Technology, 1-8-31 Midorioka, Ikeda, Osaka 563-8577, Japan, E-mail: [email protected] Abstract: G protein-coupled receptors (GPCRs) detect and distinguish between various subtypes of extracellular sig- nals, such as neurotransmitters, hormones, light, and odorous chemicals. As determinants for robust and appropriate cellular responses, common and unique features of interactions between GPCRs and their target G proteins provide insights into structure-based drug design for treatment of GPCR-related diseases. Re- cently, we found that the hydrophobic core buried between GPCR helix 8 and TM1–2 is essential for acti- vation of both specific and nonspecific G proteins. Furthermore, the 2nd residue of helix 8 is responsible for initial, transient, and specific interaction with a target G protein. Analysis of human and murine olfactory receptors (ORs) and other class-A GPCRs revealed that several amino acids, such as Glu, Gln, and Asp, are conserved at this position. -

Melatonin Mitigates the Adverse Effect of Hypoxia During Myocardial
J Vet Sci. 2021 Jul;22(4):e54 https://doi.org/10.4142/jvs.2021.22.e54 pISSN 1229-845X·eISSN 1976-555X Original Article Melatonin mitigates the adverse Molecular and Cellular Biology effect of hypoxia during myocardial differentiation in mouse embryonic stem cells Jae-Hwan Lee , Yeong-Min Yoo , Bonn Lee , SunHwa Jeong , Dinh Nam Tran , Eui-Bae Jeung * Laboratory of Veterinary Biochemistry and Molecular Biology, College of Veterinary Medicine, Chungbuk National University, Cheongju 28644, Korea Received: Mar 30, 2021 ABSTRACT Revised: Jun 7, 2021 Accepted: Jun 20, 2021 Published online: Jun 24, 2021 Background: Hypoxia causes oxidative stress and affects cardiovascular function and the programming of cardiovascular disease. Melatonin promotes antioxidant enzymes such as *Corresponding author: superoxide dismutase, glutathione reductase, glutathione peroxidase, and catalase. Eui-Bae Jeung Objectives: This study aims to investigate the correlation between melatonin and hypoxia Laboratory of Veterinary Biochemistry and Molecular Biology, College of Veterinary induction in cardiomyocytes differentiation. Medicine, Chungbuk National University, 1 Methods: Mouse embryonic stem cells (mESCs) were induced to myocardial differentiation. Chungdae-ro, Seowon-gu, Cheongju 28644, To demonstrate the influence of melatonin under hypoxia, mESC was pretreated with Korea. melatonin and then cultured in hypoxic condition. The cardiac beating ratio of the mESC- E-mail: [email protected] derived cardiomyocytes, mRNA and protein expression levels were investigated. © 2021 The Korean Society of Veterinary Results: Under hypoxic condition, the mRNA expression of cardiac-lineage markers Science (Brachyury, Tbx20, and cTn1) and melatonin receptor (Mtnr1a) was reduced. The mRNA This is an Open Access article distributed expression of cTn1 and the beating ratio of mESCs increased when melatonin was treated under the terms of the Creative Commons simultaneously with hypoxia, compared to when only exposed to hypoxia. -

Detection of Polymorphisms in the MTNR1A Gene and Their Association with Reproductive Performance in Awassi Ewes
animals Article Detection of Polymorphisms in the MTNR1A Gene and Their Association with Reproductive Performance in Awassi Ewes Giovanni Cosso 1, Michella Nehme 2, Sebastiano Luridiana 1 , Luisa Pulinas 1, Giulio Curone 3 , Chadi Hosri 4, Vincenzo Carcangiu 1 and Maria Consuelo Mura 1,* 1 Department of Veterinary Medicine of Sassari, University of Sassari, Via Vienna 2, 07100 Sassari, Italy; [email protected] (G.C.); [email protected] (S.L.); [email protected] (L.P.); [email protected] (V.C.) 2 Department of Agriculture and Food Engineering, Faculty of Engineering, Holy Spirit University of Kaslik, Kaslik, Jounieh 446, Lebanon; [email protected] 3 Department of Veterinary Medicine of Milan, University of Milan, Via dell’Università 6, 26900 Lodi, Italy; [email protected] 4 Department of Veterinary Sciences, Faculty of Agriculture, Lebanese University, Dekwaneh, Beirut 14/6573, Lebanon; [email protected] * Correspondence: [email protected]; Tel.: +39-079-229-437; Fax: +39-079-229-592 Simple Summary: The purpose of the study was to explore the influence of MTNR1A gene polymor- phisms on the reproductive performance in Awassi sheep, which is an important and widespread breed in developing Mediterranean countries. A total of 31 SNPs was detected, 5 of which caused amino acid changes. Two of the found SNPs were found to be totally linked and associated with an advanced reproductive recovery in ewes carrying the C allele. The obtained results could be useful for improving reproductive management in developing Mediterranean areas. Citation: Cosso, G.; Nehme, M.; Abstract: The economy in Mediterranean areas is tightly linked to the evolution of the sheep-farming Luridiana, S.; Pulinas, L.; Curone, G.; system; therefore, improvement in ewe’s reproductive performance is essential in the developing Hosri, C.; Carcangiu, V.; Mura, M.C. -

Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Amy Sue Bogard University of Tennessee Health Science Center
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 12-2013 Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Amy Sue Bogard University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Medical Cell Biology Commons, and the Medical Molecular Biology Commons Recommended Citation Bogard, Amy Sue , "Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells" (2013). Theses and Dissertations (ETD). Paper 330. http://dx.doi.org/10.21007/etd.cghs.2013.0029. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Document Type Dissertation Degree Name Doctor of Philosophy (PhD) Program Biomedical Sciences Track Molecular Therapeutics and Cell Signaling Research Advisor Rennolds Ostrom, Ph.D. Committee Elizabeth Fitzpatrick, Ph.D. Edwards Park, Ph.D. Steven Tavalin, Ph.D. Christopher Waters, Ph.D. DOI 10.21007/etd.cghs.2013.0029 Comments Six month embargo expired June 2014 This dissertation is available at UTHSC Digital Commons: https://dc.uthsc.edu/dissertations/330 Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells A Dissertation Presented for The Graduate Studies Council The University of Tennessee Health Science Center In Partial Fulfillment Of the Requirements for the Degree Doctor of Philosophy From The University of Tennessee By Amy Sue Bogard December 2013 Copyright © 2013 by Amy Sue Bogard. -

Seasonal Reproduction in Vertebrates: Melatonin Synthesis, Binding, and Functionality Using Tinbergen’S Four Questions
molecules Review Seasonal Reproduction in Vertebrates: Melatonin Synthesis, Binding, and Functionality Using Tinbergen’s Four Questions Dax viviD * and George E. Bentley Berkeley Department of Integrative Biology, University of California, Berkeley, CA 94720, USA; [email protected] * Correspondence: [email protected] Received: 22 December 2017; Accepted: 13 February 2018; Published: 13 March 2018 Abstract: One of the many functions of melatonin in vertebrates is seasonal reproductive timing. Longer nights in winter correspond to an extended duration of melatonin secretion. The purpose of this review is to discuss melatonin synthesis, receptor subtypes, and function in the context of seasonality across vertebrates. We conclude with Tinbergen’s Four Questions to create a comparative framework for future melatonin research in the context of seasonal reproduction. Keywords: melatonin; seasonal reproduction; vertebrates 1. Introduction “We begin in the dark, and birth is the death of us.” Antigon¯´e, Translated by Anne Carson The connection between darkness and birth is inextricable for photoperiodic, seasonally breeding animals. The tilt and rotation of the Earth creates geographical variation in temperature and daylength. Daily and annual fluctuations of environmental conditions outside of the tropics, resulting from these astronomical circumstances, correspond to changes in relative resource abundance. Organisms that detect light with the physiological mechanisms to track and store photic information over time can use daylength to anticipate environmental changes, subsequently altering behavior and physiology to optimize metabolism and seasonal reproductive timing. The synthesis of N-acetyl-5-methoxytryptamine, or melatonin, corresponds with darkness, thereby transducing photic information at the physiological level. The focus of this review is to overview melatonin research in the context of seasonal reproduction in photoperiodic breeding mammals, birds, and other vertebrates. -
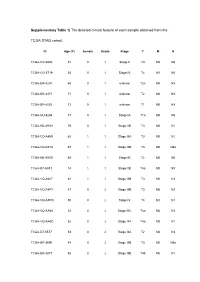
Supplementary Table 1| the Detailed Clinical Feature of Each Sample Obtained from The
Supplementary Table 1| The detailed clinical feature of each sample obtained from the TCGA STAD cohort. ID Age (Y) Gender Grade Stage T M N TCGA-CD-5800 51 0 1 Stage II T3 M0 N0 TCGA-CG-5719 54 0 1 Stage IV T4 M1 N0 TCGA-BR-4201 66 0 1 unknow T2a M0 NX TCGA-BR-4371 71 0 1 unknow T2 M0 NX TCGA-BR-4292 73 0 1 unknow T1 M0 NX TCGA-HU-8244 77 0 1 Stage IA T1a M0 N0 TCGA-KB-A93H 79 0 1 Stage IIB T3 M0 N1 TCGA-CD-A4MI 62 1 1 Stage IIIA T3 M0 N1 TCGA-VQ-A91A 67 1 1 Stage IIIB T3 M0 N3a TCGA-KB-A93G 68 1 1 Stage IB T2 M0 N0 TCGA-B7-A5TJ 74 1 1 Stage IIB T4a M0 NX TCGA-VQ-A927 81 1 1 Stage IIIB T3 M0 N3 TCGA-VQ-A8PY 47 0 2 Stage IIIB T3 M0 N2 TCGA-VQ-A8PQ 50 0 2 Stage IV T4 M1 N1 TCGA-VQ-AA68 52 0 2 Stage IIIC T4a M0 N3 TCGA-VQ-AA6D 52 0 2 Stage IIIA T4a M0 N1 TCGA-D7-5577 53 0 2 Stage IIIA T2 M0 N3 TCGA-BR-8690 54 0 2 Stage IIIB T3 M0 N3a TCGA-BR-8077 58 0 2 Stage IIIB T4b M0 N1 TCGA-HU-A4H2 58 0 2 Stage IIIB T3 M0 N3b TCGA-VQ-A91N 59 0 2 Stage IV T4 M0 N3 TCGA-D7-A74A 61 0 2 Stage IIIA T3 M0 N2 TCGA-HU-A4GP 62 0 2 Stage IIA T2 M0 N1 TCGA-BR-7716 62 0 2 Stage IIB T3 M0 N1 TCGA-BR-8679 63 0 2 Stage IB T2 M0 N0 TCGA-BR-8373 65 0 2 Stage IIIA T4a M0 N1 TCGA-VQ-A8PB 65 0 2 Stage II T3 M0 N0 TCGA-R5-A7ZF 65 0 2 Stage IV T4a M1 N1 TCGA-CG-4460 66 0 2 Stage IV T4 M1 N1 TCGA-D7-8579 66 0 2 Stage IIB T2 M0 N2 TCGA-R5-A7ZE 66 0 2 Stage IIIA T3 M0 N2 TCGA-VQ-A91Z 67 0 2 Stage IIIA T3 M0 N1 TCGA-HU-A4G9 67 0 2 Stage IA T1a M0 N0 TCGA-D7-6526 67 0 2 Stage IIIA T3 M0 N2 TCGA-BR-A4PE 68 0 2 Stage IB T2 M0 N0 TCGA-VQ-A8DL 69 0 2 Stage IIA T3 M0 N0 TCGA-BR-A4J6 -

“Genetic and Molecular Basis of Reproductive Efficiency in Swine”
UNIVERSITAT AUTÒNOMA DE BARCELONA Departament de Ciència Animal i dels Aliments Facultat de Veterinària CENTRE DE RECERCA EN AGRIGENÒMICA Departament de Genètica Animal “Genetic and molecular basis of reproductive efficiency in swine” Sarai Córdoba Terreros PhD Thesis in Animal Production Bellaterra, 2015 Supervisor: Dr. Armand Sánchez Bonastre “It is a miracle that curiosity survives formal education.” - Albert Einstein (1879-1955) A mis padres, por su incondicionalidad. El Dr. Armand Sánchez Bonastre, catedràtic del Departament de Ciència Animal i dels Aliments de la Universitat Autònoma de Barcelona, CERTIFICA: Que la Sarai Córdoba Terreros ha realitzat sota la seva direcció el treball de recerca “Genetic and molecular basis of reproductive efficiency in swine” per a obtenir el grau de Doctora per la Universitat Autònoma de Barcelona. Que aquest treball s’ha dut a terme al Departament de Ciència Animal i dels Aliments de la Facultat de Veterinària de la Universitat Autònoma de Barcelona i a la unitat de Genètica Animal del Centre de Recerca en Agrigenòmica. Bellaterra, a 9 d’Octubre de 2015 Dr. Armand Sánchez Bonastre CONTENT SUMMARY/RESUM ……………………………….………………………………………... 13 List of Tables ………………………………………………………………………… 17 List of Figures ………………………………………………………………………… 19 List of publications ………………………………………………………………………… 21 Related publications by the author ……………………………………………………… 22 Abbreviations ………………………………………………………………………… 23 1. GENERAL INTRODUCTION …………………………………………………….. 29 1.1. Pig production …………………………………………………….. 29 1.1.1. Improvement of pig production ………………………………….. 30 1.2. Reproduction in swine …………………………………………………….. 30 1.2.1. Reproductive cycle of a sow ………………………………….. 30 1.2.1.1. Follicular phase ………………………………….. 31 1.2.1.2. Luteal phase ………………………………….. 31 1.2.1.3. Pregnancy establishment ………………………… 32 1.3. Genetic basis of swine reproduction …………………………………. 33 1.3.1. -

Mapping of the Melatonin Receptor 1A (MTNR1A) Gene in Pigs, Sheep and Cattle
Mapping of the Melatonin Receptor 1a (MTNR1A) Gene in Pigs, Sheep and Cattle L. A. Messer, graduate student Chain Reaction-Restriction Fragment Length Polymorphism L. Wang, postdoctoral associate (PCR-RFLP) and Southern blot hybridization analysis to map C. K. Tuggle, professor MTNR1A in pigs, sheep and cattle. M. F. Rothschild, professor Department of Animal Science Materials and Methods PCR primers were designed from coding region of the human ASL-R1377 and sheep sequence. A 520 base pair (bp) product was purified and sequenced. This fragment was used to detect potential Summary and Implications polymorphisms on membranes containing American and Human and sheep Melatonin receptor 1a (MTNR1A) Chinese breeds and PiGMaP DNA (Archibald et al. 1995). gene information was used to clone a portion of the coding Physical mapping data was obtained by using the pig x rodent region of this gene in pigs, and to identify polymorphisms somatic cell hybrid panel (Yerle et al. 1996) and a pig YAC of the gene for its assignment to both the genetic linkage library (Rogel-Gaillard et al. 1996). This approach determines and physical maps. MTNR1A maps to pig chromosome where a gene is physically located by performing PCR 17, establishing a new region of conserved synteny experiments with whole and fragmented pig chromosomes in a between this chromosome and human chromosome 4. present/not present PCR strategy. Furthermore, we have assigned MTNR1A to bovine A portion of the sheep and cattle MTNR1A gene was chromosome 27 and sheep chromosome 26. The addition of produced from genomic DNA using primers designed from sheep genes like MTNR1A to livestock genome maps allows sequence (Reppert et al.