Evolution and Development of Shape: Integrating Quantitative Approaches
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Functional Analysis of the Homeobox Gene Tur-2 During Mouse Embryogenesis
Functional Analysis of The Homeobox Gene Tur-2 During Mouse Embryogenesis Shao Jun Tang A thesis submitted in conformity with the requirements for the Degree of Doctor of Philosophy Graduate Department of Molecular and Medical Genetics University of Toronto March, 1998 Copyright by Shao Jun Tang (1998) National Library Bibriothèque nationale du Canada Acquisitions and Acquisitions et Bibiiographic Services seMces bibliographiques 395 Wellington Street 395, rue Weifington OtbawaON K1AW OttawaON KYAON4 Canada Canada The author has granted a non- L'auteur a accordé une licence non exclusive licence alIowing the exclusive permettant à la National Library of Canada to Bibliothèque nationale du Canada de reproduce, loan, distri%uteor sell reproduire, prêter' distribuer ou copies of this thesis in microform, vendre des copies de cette thèse sous paper or electronic formats. la forme de microfiche/nlm, de reproduction sur papier ou sur format électronique. The author retains ownership of the L'auteur conserve la propriété du copyright in this thesis. Neither the droit d'auteur qui protège cette thèse. thesis nor substantial extracts fkom it Ni la thèse ni des extraits substantiels may be printed or otherwise de celle-ci ne doivent être imprimés reproduced without the author's ou autrement reproduits sans son permission. autorisation. Functional Analysis of The Homeobox Gene TLr-2 During Mouse Embryogenesis Doctor of Philosophy (1998) Shao Jun Tang Graduate Department of Moiecular and Medicd Genetics University of Toronto Abstract This thesis describes the clonhg of the TLx-2 homeobox gene, the determination of its developmental expression, the characterization of its fiuiction in mouse mesodem and penpheral nervous system (PNS) developrnent, the regulation of nx-2 expression in the early mouse embryo by BMP signalling, and the modulation of the function of nX-2 protein by the 14-3-3 signalling protein during neural development. -

Lineage-Specific Mapping of Quantitative Trait Loci
Heredity (2013) 111, 106–113 & 2013 Macmillan Publishers Limited All rights reserved 0018-067X/13 www.nature.com/hdy ORIGINAL ARTICLE Lineage-specific mapping of quantitative trait loci C Chen1 and K Ritland We present an approach for quantitative trait locus (QTL) mapping, termed as ‘lineage-specific QTL mapping’, for inferring allelic changes of QTL evolution along with branches in a phylogeny. We describe and analyze the simplest case: by adding a third taxon into the normal procedure of QTL mapping between pairs of taxa, such inferences can be made along lineages to a presumed common ancestor. Although comparisons of QTL maps among species can identify homology of QTLs by apparent co- location, lineage-specific mapping of QTL can classify homology into (1) orthology (shared origin of QTL) versus (2) paralogy (independent origin of QTL within resolution of map distance). In this light, we present a graphical method that identifies six modes of QTL evolution in a three taxon comparison. We then apply our model to map lineage-specific QTLs for inbreeding among three taxa of yellow monkey-flower: Mimulus guttatus and two inbreeders M. platycalyx and M. micranthus, but critically assuming outcrossing was the ancestral state. The two most common modes of homology across traits were orthologous (shared ancestry of mutation for QTL alleles). The outbreeder M. guttatus had the fewest lineage-specific QTL, in accordance with the presumed ancestry of outbreeding. Extensions of lineage-specific QTL mapping to other types of data and crosses, and to inference of ancestral QTL state, are discussed. Heredity (2013) 111, 106–113; doi:10.1038/hdy.2013.24; published online 24 April 2013 Keywords: Quantitative trait locus mapping; evolution of mating systems; Mimulus INTRODUCTION common ancestor of these two taxa, back to the common ancestor of Knowledge of the genetic architecture of complex traits can offer all three taxa and forwards again to the third taxon. -
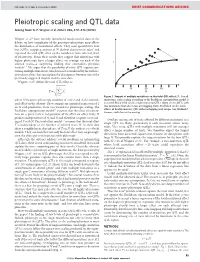
Pleiotropic Scaling and QTL Data Arising From: G
NATURE | Vol 456 | 4 December 2008 BRIEF COMMUNICATIONS ARISING Pleiotropic scaling and QTL data Arising from: G. P. Wagner et al. Nature 452, 470–473 (2008) Wagner et al.1 have recently introduced much-needed data to the debate on how complexity of the genotype–phenotype map affects the distribution of mutational effects. They used quantitative trait loci (QTLs) mapping analysis of 70 skeletal characters in mice2 and regressed the total QTL effect on the number of traits affected (level of pleiotropy). From their results they suggest that mutations with higher pleiotropy have a larger effect, on average, on each of the T affected traits—a surprising finding that contradicts previous models3–7. We argue that the possibility of some QTL regions con- taining multiple mutations, which was not considered by the authors, introduces a bias that can explain the discrepancy between one of the previously suggested models and the new data. Wagner et al.1 define the total QTL effect as vffiffiffiffiffiffiffiffiffiffiffiffiffi u uXN 10 20 30 ~t 2 T Ai N i~1 Figure 1 | Impact of multiple mutations on the total QTL effect, T. For all where N measures pleiotropy (number of traits) and Ai the normal- mutations, strict scaling according to the Euclidean superposition model is ized effect on the ith trait. They compare an empirical regression of T assumed. Black filled circles, single-mutation QTLs. Open circles, QTLs with on N with predictions from two models for pleiotropic scaling. The two mutations that affect non-overlapping traits. Red filled circles, total Euclidean superposition model3,4 assumes that the effect of a muta- effects of double-mutant QTL with overlapping trait ranges (see Methods). -

Products for Morphogen Research
R&D Systems Tools for Cell Biology Research™ Products for Morphogen Research BMP-4 NEURAL PLATE BMP-7 PROSPECTIVE NEURAL CREST NON-NEURAL ECTODERM Noggin Shh Noggin PRESOMITIC MESODERM NOTOCHORD NON-NEURAL ECTODERM FUTURE FLOOR PLATE BMP-4 Shh Noggin DORSAL AORTA ROOF PLATE Products for Morphogen Research for Products Noggin Wnt-1 Wnt-3a Wnt-4 NT-3 Wnt-6 Wnt-7a EARLY SOMITE Myf5 NEURAL TUBE Pax3 Sim1 BMP-4 INTERMEDIATE MESODERM Shh ShhShh NogginNoggin BMP-4 DORSAL AORTA Ihh NOTOCHORD MORPHOGENS Morphogens are molecules that regulate cell fate during development. Formation of morphogen concentration gradients directs the biological responses of surrounding cells. Graded responses occur as a result of morphogens binding to specific cell surface receptors that subsequently activate intracellular signaling pathways and promote or repress gene expression at specific threshold concentrations. Activation or inactivation of these signaling pathways provides positional information that ultimately determines tissue organization and morphology. Research in model organisms has revealed that morphogens are involved in many aspects of development. For example, morphogens are required in Drosophila for patterning of the dorso-ventral and anterior-posterior axes, segment patterning, and positional signaling in the leg and wing imaginal discs. Proteins belonging to the Wingless/Wnt, Notch, Hedgehog, and TGF-b families have been identified as morphogens that direct a number of these processes. Research in higher organisms has demonstrated that homologues of these same signaling molecules regulate vertebrate axis formation, anterior/posterior polarity during limb development, mesoderm patterning, and numerous other processes that establish an organism’s basic body structure. R&D Systems offers a wide selection of proteins, antibodies, and ELISAs for morphogen-related developmental research. -

1 ROBERT PLOMIN on Behavioral Genetics Genetic Links Common
Genetic links • Background – Why has educational psychology ben so slow ROBERT PLOMIN to accept genetics? on Behavioral Genetics • Beyond nature vs. nurture – 3 findings from genetic research with far- reaching implications for education: October, 2004 The nature of nurture The abnormal is normal Generalist genes • Molecular Genetics Quantitative genetics: twin and adoption studies Common medical disorders Identical twins Fraternal twins (monozygotic, MZ) (dizygotic, DZ) 1 Twin concordances in TEDS at 7 years Twins Early Development Study (TEDS) for low reading and maths Perceptions of nature/nurture: % parents (N=1340) and teachers Beyond nature vs. nurture: (N=667) indicating that genes are at least as important as environment (Walker & Plomin, Ed. Psych., in press) The nature of nurture • Genetic influence on experience • Genotype-environment correlation – Children’s genetic propensities are correlated with their experiences passive evocative active 2 Beyond nature vs. nurture: Genotype-environment correlation The nature of nurture • Passive • Genotype-environment correlation – e.g., family background (SES) and school – Children’s genetic propensities are correlated with achievement their experiences passive • Evocative evocative – Children evoke experiences for genetic reasons Active • Active • Molecular genetics – Children actively create environments that foster – Find genes associated with learning experiences their genetic propensities Beyond nature vs. nurture: The one-gene one-disability (OGOD) model The abnormal is normal • Common learning disabilities are the quantitative extreme of the same genetic and environmental factors responsible for normal variation in learning abilities. • There are no common learning disabilities, just the extremes of normal variation. 3 OGOD (rare) and QTL (common) disorders The quantitative trait locus (QTL) model Beyond nature vs. -

Rapid Changes in Morphogen Concentration Control Self-Organized
RESEARCH COMMUNICATION Rapid changes in morphogen concentration control self-organized patterning in human embryonic stem cells Idse Heemskerk1†, Kari Burt1, Matthew Miller1, Sapna Chhabra2, M Cecilia Guerra1, Lizhong Liu1, Aryeh Warmflash1,3* 1Department of Biosciences, Rice University, Houston, United States; 2Systems, Synthetic and Physical Biology Program, Rice University, Houston, United States; 3Department of Bioengineering, Rice University, Houston, United States Abstract During embryonic development, diffusible signaling molecules called morphogens are thought to determine cell fates in a concentration-dependent way. Yet, in mammalian embryos, concentrations change rapidly compared to the time for making cell fate decisions. Here, we use human embryonic stem cells (hESCs) to address how changing morphogen levels influence differentiation, focusing on how BMP4 and Nodal signaling govern the cell-fate decisions associated with gastrulation. We show that BMP4 response is concentration dependent, but that expression of many Nodal targets depends on rate of concentration change. Moreover, in a self- organized stem cell model for human gastrulation, expression of these genes follows rapid changes in endogenous Nodal signaling. Our study shows a striking contrast between the specific ways ligand dynamics are interpreted by two closely related signaling pathways, highlighting both the *For correspondence: subtlety and importance of morphogen dynamics for understanding mammalian embryogenesis [email protected] and designing optimized protocols for directed stem cell differentiation. Editorial note: This article has been through an editorial process in which the authors decide how † Present address: Department to respond to the issues raised during peer review. The Reviewing Editor’s assessment is that all of Cell and Developmental the issues have been addressed (see decision letter). -

Annominer Is a New Web-Tool to Integrate Epigenetics, Transcription
www.nature.com/scientificreports OPEN AnnoMiner is a new web‑tool to integrate epigenetics, transcription factor occupancy and transcriptomics data to predict transcriptional regulators Arno Meiler1,3, Fabio Marchiano2,3, Margaux Haering2, Manuela Weitkunat1, Frank Schnorrer1,2 & Bianca H. Habermann1,2* Gene expression regulation requires precise transcriptional programs, led by transcription factors in combination with epigenetic events. Recent advances in epigenomic and transcriptomic techniques provided insight into diferent gene regulation mechanisms. However, to date it remains challenging to understand how combinations of transcription factors together with epigenetic events control cell‑type specifc gene expression. We have developed the AnnoMiner web‑server, an innovative and fexible tool to annotate and integrate epigenetic, and transcription factor occupancy data. First, AnnoMiner annotates user‑provided peaks with gene features. Second, AnnoMiner can integrate genome binding data from two diferent transcriptional regulators together with gene features. Third, AnnoMiner ofers to explore the transcriptional deregulation of genes nearby, or within a specifed genomic region surrounding a user‑provided peak. AnnoMiner’s fourth function performs transcription factor or histone modifcation enrichment analysis for user‑provided gene lists by utilizing hundreds of public, high‑quality datasets from ENCODE for the model organisms human, mouse, Drosophila and C. elegans. Thus, AnnoMiner can predict transcriptional regulators for a studied -

Epigenome-Wide Study Identified Methylation Sites Associated With
nutrients Article Epigenome-Wide Study Identified Methylation Sites Associated with the Risk of Obesity Majid Nikpay 1,*, Sepehr Ravati 2, Robert Dent 3 and Ruth McPherson 1,4,* 1 Ruddy Canadian Cardiovascular Genetics Centre, University of Ottawa Heart Institute, 40 Ruskin St–H4208, Ottawa, ON K1Y 4W7, Canada 2 Plastenor Technologies Company, Montreal, QC H2P 2G4, Canada; [email protected] 3 Department of Medicine, Division of Endocrinology, University of Ottawa, the Ottawa Hospital, Ottawa, ON K1Y 4E9, Canada; [email protected] 4 Atherogenomics Laboratory, University of Ottawa Heart Institute, Ottawa, ON K1Y 4W7, Canada * Correspondence: [email protected] (M.N.); [email protected] (R.M.) Abstract: Here, we performed a genome-wide search for methylation sites that contribute to the risk of obesity. We integrated methylation quantitative trait locus (mQTL) data with BMI GWAS information through a SNP-based multiomics approach to identify genomic regions where mQTLs for a methylation site co-localize with obesity risk SNPs. We then tested whether the identified site contributed to BMI through Mendelian randomization. We identified multiple methylation sites causally contributing to the risk of obesity. We validated these findings through a replication stage. By integrating expression quantitative trait locus (eQTL) data, we noted that lower methylation at cg21178254 site upstream of CCNL1 contributes to obesity by increasing the expression of this gene. Higher methylation at cg02814054 increases the risk of obesity by lowering the expression of MAST3, whereas lower methylation at cg06028605 contributes to obesity by decreasing the expression of SLC5A11. Finally, we noted that rare variants within 2p23.3 impact obesity by making the cg01884057 Citation: Nikpay, M.; Ravati, S.; Dent, R.; McPherson, R. -

QUANTITATIVE TRAIT LOCUS ANALYSIS: in Several Variants (I.E., Alleles)
TECHNOLOGIES FROM THE FIELD QUANTITATIVE TRAIT LOCUS ANALYSIS: in several variants (i.e., alleles). QTL analysis allows one MULTIPLE CROSS AND HETEROGENEOUS to map, with some precision, the genomic position of STOCK MAPPING these alleles. For many researchers in the alcohol field, the break Robert Hitzemann, Ph.D.; John K. Belknap, Ph.D.; through with respect to the genetic determination of alcohol- and Shannon K. McWeeney, Ph.D. related behaviors occurred when Plomin and colleagues (1991) made the seminal observation that a specific panel of RI mice (i.e., the BXD panel) could be used to identify KEY WORDS: Genetic theory of alcohol and other drug use; genetic the physical location of (i.e., to map) QTLs for behavioral factors; environmental factors; behavioral phenotype; behavioral phenotypes. Because the phenotypes of the different strains trait; quantitative trait gene (QTG); inbred animal strains; in this panel had been determined for many alcohol- recombinant inbred (RI) mouse strains; quantitative traits; related traits, researchers could readily apply the strategy quantitative trait locus (QTL) mapping; multiple cross mapping of RI–QTL mapping (Gora-Maslak et al. 1991). Although (MCM); heterogeneous stock (HS) mapping; microsatellite investigators recognized early on that this panel was not mapping; animal models extensive enough to answer all questions, the emerging data illustrated the rich genetic complexity of alcohol- related phenotypes (Belknap 1992; Plomin and McClearn ntil well into the 1990s, both preclinical and clinical 1993). research focused on finding “the” gene for human The next advance came with the development of diseases, including alcoholism. This focus was rein U microsatellite maps (Dietrich et al. -

Article the Power to Detect Quantitative Trait Loci Using
The Power to Detect Quantitative Trait Loci Using Resequenced, Experimentally Evolved Populations of Diploid, Sexual Organisms James G. Baldwin-Brown,*,1 Anthony D. Long,1 and Kevin R. Thornton1 1Department of Ecology and Evolutionary Biology, University of California, Irvine *Corresponding author: E-mail: [email protected]. Associate editor: John Parsch Abstract A novel approach for dissecting complex traits is to experimentally evolve laboratory populations under a controlled environment shift, resequence the resulting populations, and identify single nucleotide polymorphisms (SNPs) and/or genomic regions highly diverged in allele frequency. To better understand the power and localization ability of such an evolve and resequence (E&R) approach, we carried out forward-in-time population genetics simulations of 1 Mb genomic regions under a large combination of experimental conditions, then attempted to detect significantly diverged SNPs. Our analysis indicates that the ability to detect differentiation between populations is primarily affected by selection coef- ficient, population size, number of replicate populations, and number of founding haplotypes. We estimate that E&R studies can detect and localize causative sites with 80% success or greater when the number of founder haplotypes is over 500, experimental populations are replicated at least 25-fold, population size is at least 1,000 diploid individuals, and the selection coefficient on the locus of interest is at least 0.1. More achievable experimental designs (less replicated, fewer founder haplotypes, smaller effective population size, and smaller selection coefficients) can have power of greater than 50% to identify a handful of SNPs of which one is likely causative. Similarly, in cases where s 0.2, less demanding experimental designs can yield high power. -

Glossary/Index
Glossary 03/08/2004 9:58 AM Page 119 GLOSSARY/INDEX The numbers after each term represent the chapter in which it first appears. additive 2 allele 2 When an allele’s contribution to the variation in a One of two or more alternative forms of a gene; a single phenotype is separately measurable; the independent allele for each gene is inherited separately from each effects of alleles “add up.” Antonym of nonadditive. parent. ADHD/ADD 6 Alzheimer’s disease 5 Attention Deficit Hyperactivity Disorder/Attention A medical disorder causing the loss of memory, rea- Deficit Disorder. Neurobehavioral disorders character- soning, and language abilities. Protein residues called ized by an attention span or ability to concentrate that is plaques and tangles build up and interfere with brain less than expected for a person's age. With ADHD, there function. This disorder usually first appears in persons also is age-inappropriate hyperactivity, impulsive over age sixty-five. Compare to early-onset Alzheimer’s. behavior or lack of inhibition. There are several types of ADHD: a predominantly inattentive subtype, a predomi- amino acids 2 nantly hyperactive-impulsive subtype, and a combined Molecules that are combined to form proteins. subtype. The condition can be cognitive alone or both The sequence of amino acids in a protein, and hence pro- cognitive and behavioral. tein function, is determined by the genetic code. adoption study 4 amnesia 5 A type of research focused on families that include one Loss of memory, temporary or permanent, that can result or more children raised by persons other than their from brain injury, illness, or trauma. -

Analysis on Gene Modular Network Reveals Morphogen-Directed Development Robustness in Drosophila
Zhang et al. Cell Discovery (2020) 6:43 Cell Discovery https://doi.org/10.1038/s41421-020-0173-z www.nature.com/celldisc ARTICLE Open Access Analysis on gene modular network reveals morphogen-directed development robustness in Drosophila Shuo Zhang1,2,JuanZhao1,3, Xiangdong Lv1,2, Jialin Fan1,2,YiLu1,TaoZeng 1,3, Hailong Wu1, Luonan Chen 1,3,4,5 and Yun Zhao1,4,6 Abstract Genetic robustness is an important characteristic to tolerate genetic or nongenetic perturbations and ensure phenotypic stability. Morphogens, a type of evolutionarily conserved diffusible molecules, govern tissue patterns in a direction-dependent or concentration-dependent manner by differentially regulating downstream gene expression. However, whether the morphogen-directed gene regulatory network possesses genetic robustness remains elusive. In the present study, we collected 4217 morphogen-responsive genes along A-P axis of Drosophila wing discs from the RNA-seq data, and clustered them into 12 modules. By applying mathematical model to the measured data, we constructed a gene modular network (GMN) to decipher the module regulatory interactions and robustness in morphogen-directed development. The computational analyses on asymptotical dynamics of this GMN demonstrated that this morphogen-directed GMN is robust to tolerate a majority of genetic perturbations, which has been further validated by biological experiments. Furthermore, besides the genetic alterations, we further demonstrated that this morphogen-directed GMN can well tolerate nongenetic perturbations (Hh production changes) via computational fi 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; analyses and experimental validation. Therefore, these ndings clearly indicate that the morphogen-directed GMN is robust in response to perturbations and is important for Drosophila to ensure the proper tissue patterning in wing disc.