Legionnaires' Disease
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Legionnaires' Disease, Pontiac Fever, Legionellosis and Legionella
Legionnaires’ Disease, Pontiac Fever, Legionellosis and Legionella Q: What is Legionellosis and who is at risk? Legionellosis is an infection caused by Legionella bacteria. Legionellosis can present as two distinct illnesses: Pontiac fever (a self-limited flu-like mild respiratory illness), and Legionnaires’ Disease (a more severe illness involving pneumonia). People of any age can get Legionellosis, but the disease occurs most frequently in persons over 50 years of age. The disease most often affects those who smoke heavily, have chronic lung disease, or have underlying medical conditions that lower their immune system, such as diabetes, cancer, or renal dysfunction. Persons taking certain drugs that lower their immune system, such as steroids, have an increased risk of being affected by Legionellosis. Many people may be infected with Legionella bacteria without developing any symptoms, and others may be treated without having to be hospitalized. Q: What is Legionella? Legionella bacteria are found naturally in freshwater environments such as creeks, ponds and lakes, as well as manmade structures such as plumbing systems and cooling towers. Legionella can multiply in warm water (77°F to 113°F). Legionella pneumophila is responsible for over 90 percent of Legionnaires’ Disease cases and several different species of Legionella are responsible for Pontiac Fever. Q: How is Legionella spread and how does someone acquire Legionellosis (Legionnaires’ Disease/Pontiac Fever)? Legionella bacteria become a health concern when they grow and spread in manmade structures such as plumbing systems, hot water tanks, cooling towers, hot tubs, decorative fountains, showers and faucets. Legionellosis is acquired after inhaling mists from a contaminated water source containing Legionella bacteria. -

NCTC) Bacterial Strain Equivalents to American Type Culture Collection (ATCC) Bacterial Strains
This list shows National Collection of Type Cultures (NCTC) bacterial strain equivalents to American Type Culture Collection (ATCC) bacterial strains. NCTC Number CurrentName ATCC Number NCTC 7212 Acetobacter pasteurianus ATCC 23761 NCTC 10138 Acholeplasma axanthum ATCC 25176 NCTC 10171 Acholeplasma equifetale ATCC 29724 NCTC 10128 Acholeplasma granularum ATCC 19168 NCTC 10172 Acholeplasma hippikon ATCC 29725 NCTC 10116 Acholeplasma laidlawii ATCC 23206 NCTC 10134 Acholeplasma modicum ATCC 29102 NCTC 10188 Acholeplasma morum ATCC 33211 NCTC 10150 Acholeplasma oculi ATCC 27350 NCTC 10198 Acholeplasma parvum ATCC 29892 NCTC 8582 Achromobacter denitrificans ATCC 15173 NCTC 10309 Achromobacter metalcaligenes ATCC 17910 NCTC 10807 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 27061 NCTC 10808 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 17062 NCTC 10809 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 27063 NCTC 12156 Acinetobacter baumannii ATCC 19606 NCTC 10303 Acinetobacter baumannii ATCC 17904 NCTC 7844 Acinetobacter calcoaceticus ATCC 15308 NCTC 12983 Acinetobacter calcoaceticus ATCC 23055 NCTC 8102 acinetobacter dna group 13 ATCC 17903 NCTC 10304 Acinetobacter genospecies 13 ATCC 17905 NCTC 10306 Acinetobacter haemolyticus ATCC 17907 NCTC 10305 Acinetobacter haemolyticus subsp haemolyticus ATCC 17906 NCTC 10308 Acinetobacter johnsonii ATCC 17909 NCTC 10307 Acinetobacter junii ATCC 17908 NCTC 5866 Acinetobacter lwoffii ATCC 15309 NCTC 12870 Actinobacillus delphinicola ATCC 700179 NCTC 8529 Actinobacillus equuli ATCC 19392 -

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

Management Weaknesses Delayed Response to Flint Water Crisis
U.S. ENVIRONMENTAL PROTECTION AGENCY OFFICE OF INSPECTOR GENERAL Ensuring clean and safe water Compliance with the law Management Weaknesses Delayed Response to Flint Water Crisis Report No. 18-P-0221 July 19, 2018 Report Contributors: Stacey Banks Charles Brunton Kathlene Butler Allison Dutton Tiffine Johnson-Davis Fred Light Jayne Lilienfeld-Jones Tim Roach Luke Stolz Danielle Tesch Khadija Walker Abbreviations CCT Corrosion Control Treatment CFR Code of Federal Regulations EPA U.S. Environmental Protection Agency FY Fiscal Year GAO U.S. Government Accountability Office LCR Lead and Copper Rule MDEQ Michigan Department of Environmental Quality OECA Office of Enforcement and Compliance Assurance OIG Office of Inspector General OW Office of Water ppb parts per billion PQL Practical Quantitation Limit PWSS Public Water System Supervision SDWA Safe Drinking Water Act Cover Photo: EPA Region 5 emergency response vehicle in Flint, Michigan. (EPA photo) Are you aware of fraud, waste or abuse in an EPA Office of Inspector General EPA program? 1200 Pennsylvania Avenue, NW (2410T) Washington, DC 20460 EPA Inspector General Hotline (202) 566-2391 1200 Pennsylvania Avenue, NW (2431T) www.epa.gov/oig Washington, DC 20460 (888) 546-8740 (202) 566-2599 (fax) [email protected] Subscribe to our Email Updates Follow us on Twitter @EPAoig Learn more about our OIG Hotline. Send us your Project Suggestions U.S. Environmental Protection Agency 18-P-0221 Office of Inspector General July 19, 2018 At a Glance Why We Did This Project Management Weaknesses -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -

Host-Adaptation in Legionellales Is 2.4 Ga, Coincident with Eukaryogenesis
bioRxiv preprint doi: https://doi.org/10.1101/852004; this version posted February 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Host-adaptation in Legionellales is 2.4 Ga, 2 coincident with eukaryogenesis 3 4 5 Eric Hugoson1,2, Tea Ammunét1 †, and Lionel Guy1* 6 7 1 Department of Medical Biochemistry and Microbiology, Science for Life Laboratories, 8 Uppsala University, Box 582, 75123 Uppsala, Sweden 9 2 Department of Microbial Population Biology, Max Planck Institute for Evolutionary 10 Biology, D-24306 Plön, Germany 11 † current address: Medical Bioinformatics Centre, Turku Bioscience, University of Turku, 12 Tykistökatu 6A, 20520 Turku, Finland 13 * corresponding author 14 1 bioRxiv preprint doi: https://doi.org/10.1101/852004; this version posted February 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 15 Abstract 16 Bacteria adapting to living in a host cell caused the most salient events in the evolution of 17 eukaryotes, namely the seminal fusion with an archaeon 1, and the emergence of both the 18 mitochondrion and the chloroplast 2. A bacterial clade that may hold the key to understanding 19 these events is the deep-branching gammaproteobacterial order Legionellales – containing 20 among others Coxiella and Legionella – of which all known members grow inside eukaryotic 21 cells 3. -

Electronic Laboratory Reporting Use Case January 2011
ILLINOIS HEALTH INFORMATION EXCHANGE Electronic Laboratory Reporting Use Case Electronic Laboratory Reporting and Health Information Exchange Illinois Health Information Exchange Public Health Work Group January 2011 Electronic Laboratory Reporting Use Case January 2011 Table of Contents 1.0 Executive Summary……………………………………………………………………….3 2.0 Introduction…………………………………………………………………...……………..5 3.0 Scope……………………………………..………………………………………………………5 4.0 Use Case Stakeholders…………………………………………………………….….....6 5.0 Issues and Obstacles……………………………………………………………………...8 6.0 Use Case Pre-Conditions .………………….…………………………………………...8 7.0 Use Case Post-Conditions.……………………………………………………………...9 8.0 Detailed Scenarios/Technical Specifications.………………………………10 9.0 Validation and Certification………………………………………………………...12 Appendix ………………………………………………………………………………………….....13 Page 2 Electronic Laboratory Reporting Use Case January 2011 1.0 Executive Summary This Use Case is a product of the Public Health Work Group (PHWG) of the Illinois Health Information Exchange (HIE) Advisory Committee. The Illinois HIE Advisory Committee was constituted as the diverse public healthcare stakeholder body providing input and recommendations on the creation of the Illinois Health Information Exchange Authority (“the Authority”) as the Illinois vehicle for designing and implementing electronic health information exchange in Illinois. The establishment of the Authority marks the formal transition of the work of the HIE Advisory Committee and the Work Groups into alignment with the provisions of Illinois -

Legionella Information for Community Public Water Systems, Health Care
Legionella Information FOR COMMUNITY PUBLIC WATER SYSTEMS (CPWSs), HEALTH CARE FACILITIES, AND ALL TYPES OF BUILDINGS What is Legionella? Legionella is a bacterium commonly found in natural and man-made aquatic environments. Legionella can be found at low concentrations in any public water system. Legionella only poses a health risk when growth occurs in warm stagnant water, the water is aerosolized, and the small droplets are inhaled. Legionella generally does not pose a health risk if a person drinks the water. Those that are infected may develop legionellosis, a type of pneumonia called Legionnaires’ disease, or a flu-like illness called Pontiac fever. There has been an increase in Legionnaires’ disease cases nationwide and in Minnesota, with 17 confirmed cases in 2004 and 115 cases in 2016. The risk to people depends on many factors, including: 1. Amplification or growth of Legionella. ▪ Conditions such as water temperatures between 77oF and 108oF, stagnation, and the presence of scale, sediment, biofilm, and protozoa promote amplification of Legionella. 2. Aerosolization of colonized water. ▪ Common modes of aerosolization include showers and faucets, cooling towers, hot tubs, and decorative fountains and water features. 3. Size of the water droplets. 4. Strain of Legionella. 5. Susceptibility of exposed individual. ▪ Those who are over 50 years of age, are male, smoke, have chronic lung disease, and/or are immunocompromised or immunosuppressed are more susceptible. According to the Centers for Disease Control and Prevention (CDC), about 5,000 cases of Legionnaires’ disease are reported each year in the United States, and one out of every 10 people who get Legionnaires’ disease will die. -

The Role of the Intensive Care Unit Environment in the Pathogenesis and Prevention of Ventilator-Associated Pneumonia
The Role of the Intensive Care Unit Environment in the Pathogenesis and Prevention of Ventilator-Associated Pneumonia Christopher J Crnich MD MSc, Nasia Safdar MD MSc, and Dennis G Maki MD Introduction Epidemiology and Pathogenesis Aerodigestive Colonization Direct Inoculation of the Lower Airway Importance of Endogenous Versus Exogenous Colonization Environmental Sources of Colonization Animate Environment Inanimate Environment The Need for a More Comprehensive Approach to the Prevention of VAP That Acknowledges the Important Role of the Hospital Environment Organizational Structure Interventions at the Interface Between the Health Care Worker and the Patient or Inanimate Environment Prevention of Infections Caused by Respiratory Devices Prevention of Infections Caused by Legionella Species Prevention of Infections Caused by Filamentous Fungi Summary Ventilator-associated pneumonia is preceded by lower-respiratory-tract colonization by pathogenic microorganisms that derive from endogenous or exogenous sources. Most ventilator-associated pneumonias are the result of exogenous nosocomial colonization, especially, pneumonias caused by resistant bacteria, such as methicillin-resistant Staphylococcus aureus and multi-resistant Acineto- bacter baumannii and Pseudomonas aeruginosa,orbyLegionella species or filamentous fungi, such as Aspergillus. Exogenous colonization originates from a very wide variety of animate and inani- mate sources in the intensive care unit environment. As a result, a strategic approach that combines measures to prevent cross-colonization with those that focus on oral hygiene and prevention of microaspiration of colonized oropharyngeal secretions should bring the greatest reduction in the risk of ventilator-associated pneumonia. This review examines strategies to prevent transmission of environmental pathogens to the vulnerable mechanically-ventilated patient. Key words: ventilator- associated pneumonia. [Respir Care 2005;50(6):813–836. -

Legionella Monitoring in NYC's Distribution System
Legionella Monitoring in NYC’s Distribution System ENOMA OMOREGIE NEW YORK CITY DEPARTMENT OF HEALTH AND MENTAL HYGIENE PUBLIC HEALTH LABORATORY NATIONAL ENVIRONMENTAL MONITORING CONFERENCE 2019 Legionella spp. Monitoring in the New York City Drinking Water Distribution System (LMINDS) Joint study between the NYC Department of Environmental Protection and Department of Health and Mental Hygiene. Four primary study questions: What is the prevalence and geographic distribution of Legionella in NYC’s water distribution system? What Legionella species inhabit the NYC distribution system, and how do they relate to species described in clinical and environmental studies? How do environmental factors such as physiochemical water parameters, water age, disinfectant levels etc., and utility management practices correlate with the presence of Legionella? Can utility management practices that reduce Legionella levels be identified? What is Legionella? Gram negative bacterium that is part of the Gammaproteobacteria. Naturally occurring and often found in aquatic systems: rivers, streams, cooling towers, hot and cold domestic water systems etc. Grows optimally at around 35°C. Causative agent of legionellosis. Source: CDC.gov; Chemology.com.au Legionellosis is two distinct syndromes Legionnaires’ disease, which is a serious type of pneumonia. ~6,000 reported cases per year in the USA and NYC has about 200 ‐ 440 cases per year. 9% mortality rate. Risk factors include smoking, chronic lung disease, age, and a compromised immune system. -
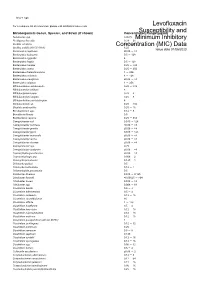
Susceptibility and Resistance Data
toku-e logo For a complete list of references, please visit antibiotics.toku-e.com Levofloxacin Microorganism Genus, Species, and Strain (if shown) Concentration Range (μg/ml)Susceptibility and Aeromonas spp. 0.0625 Minimum Inhibitory Alcaligenes faecalis 0.39 - 25 Bacillus circulans Concentration0.25 - 8 (MIC) Data Bacillus subtilis (ATCC 6051) 6.25 Issue date 01/06/2020 Bacteroides capillosus ≤0.06 - >8 Bacteroides distasonis 0.5 - 128 Bacteroides eggerthii 4 Bacteroides fragilis 0.5 - 128 Bacteroides merdae 0.25 - >32 Bacteroides ovatus 0.25 - 256 Bacteroides thetaiotaomicron 1 - 256 Bacteroides uniformis 4 - 128 Bacteroides ureolyticus ≤0.06 - >8 Bacteroides vulgatus 1 - 256 Bifidobacterium adolescentis 0.25 - >32 Bifidobacterium bifidum 8 Bifidobacterium breve 0.25 - 8 Bifidobacterium longum 0.25 - 8 Bifidobacterium pseudolongum 8 Bifidobacterium sp. 0.25 - >32 Bilophila wadsworthia 0.25 - 16 Brevibacterium spp. 0.12 - 8 Brucella melitensis 0.5 Burkholderia cepacia 0.25 - 512 Campylobacter coli 0.015 - 128 Campylobacter concisus ≤0.06 - >8 Campylobacter gracilis ≤0.06 - >8 Campylobacter jejuni 0.015 - 128 Campylobacter mucosalis ≤0.06 - >8 Campylobacter rectus ≤0.06 - >8 Campylobacter showae ≤0.06 - >8 Campylobacter spp. 0.25 Campylobacter sputorum ≤0.06 - >8 Capnocytophaga ochracea ≤0.06 - >8 Capnocytophaga spp. 0.006 - 2 Chlamydia pneumonia 0.125 - 1 Chlamydia psittaci 0.5 Chlamydia trachomatis 0.12 - 1 Chlamydophila pneumonia 0.5 Citrobacter diversus 0.015 - 0.125 Citrobacter freundii ≤0.00625 - >64 Citrobacter koseri 0.015 - -

Aquascreen® Legionella Species Qpcr Detection Kit
AquaScreen® Legionella species qPCR Detection Kit INSTRUCTIONS FOR USE FOR USE IN RESEARCH AND QUALITY CONTROL Symbols Lot No. Cat. No. Expiry date Storage temperature Number of reactions Manufacturer INDICATION The AquaScreen® Legionella species qPCR Detection kit is specifically designed for the quantitative detection of several Legionella species in water samples prepared with the AquaScreen® FastExt- ract kit. Its design complies with the requirements of AFNOR T90-471 and ISO/TS 12869:2012. Legionella are ubiquitous bacteria in surface water and moist soil, where they parasitize protozoa. The optimal growth temperature lies between +15 and +45 °C, whereas these gram-negative bacteria are dormant below 20 °C and do not survive above 60 °C. Importantly, Legionella are well-known as opportunistic intracellular human pathogens causing Legionnaires’ disease and Pontiac fever. The transmission occurs through inhalation of contami- nated aerosols generated by an infected source (e.g. human-made water systems like shower- heads, sink faucets, heaters, cooling towers, and many more). In order to efficiently prevent Legionella outbreaks, water safety control measures need syste- matic application but also reliable validation by fast Legionella testing. TEST PRINCIPLE The AquaScreen® Legionella species Kit uses qPCR for quantitative detection of legionella in wa- ter samples. In contrast to more time-consuming culture-based methods, AquaScreen® assays need less than six hours including sample preparation and qPCR to reliably detect Legionella. Moreover, the AquaScreen® qPCR assay has proven excellent performance in terms of specificity and sensitivity: other bacterial genera remain undetected whereas linear quantification is obtai- ned up to 1 x 106 particles per sample, therefore requiring no material dilution.