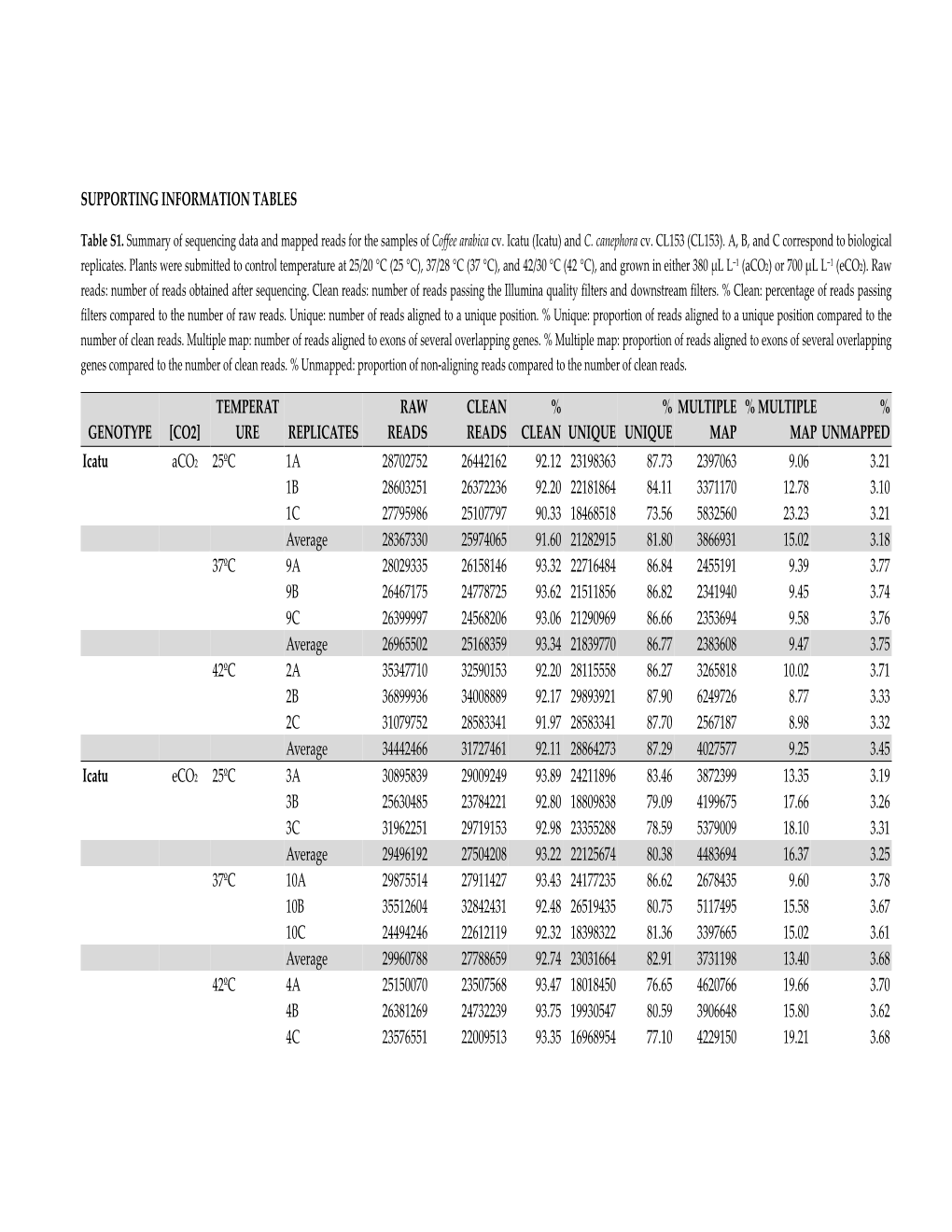
Supporting Information Tables Genotype
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

METACYC ID Description A0AR23 GO:0004842 (Ubiquitin-Protein Ligase
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Heat Stress Responsive Zostera marina Genes, Southern Population (α=0. -

I HIGH MASS ACCURACY COUPLED to SPATIALLY-DIRECTED
HIGH MASS ACCURACY COUPLED TO SPATIALLY-DIRECTED PROTEOMICS FOR IMPROVED PROTEIN IDENTIFICATIONS IN IMAGING MASS SPECTROMETRY EXPERIMENTS By David Geoffrey Rizzo Dissertation Submitted to the Faculty of the Graduate School of Vanderbilt University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in Chemistry August, 2016 Nashville, Tennessee Approved: Richard M. Caprioli, Ph.D. Kevin L. Schey, Ph.D. John A. McLean, Ph.D. Michael P. Stone, Ph.D. i Copyright © 2016 by David Geoffrey Rizzo All Rights Reserved ii This work is dedicated to my family and friends, who have shown nothing but support for me in all of life’s endeavors. iii ACKNOWLEDGEMENTS “As we express our gratitude, we must never forget that the highest appreciation is not to utter words, but to live by them.” - John F. Kennedy – There are many people I must thank for showing kindness, encouragement, and support for me during my tenure as a graduate student. First and foremost, I would like to thank my research advisor, Richard Caprioli, for providing both ample resources and guidance that allowed me to grow as a scientist. Our discussions about my research and science in general have helped me become a much more focused and discerning analytical chemist. I must also thank my Ph.D. committee members, Drs. Kevin Schey, John McLean, and Michael Stone, who have brought valuable insight into my research and provided direction along the way. My undergraduate advisor, Dr. Facundo Fernández, encouraged me to begin research in his lab and introduced me to the world of mass spectrometry. -

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Light-Induced Psba Translation in Plants Is Triggered by Photosystem II Damage Via an Assembly-Linked Autoregulatory Circuit
Light-induced psbA translation in plants is triggered by photosystem II damage via an assembly-linked autoregulatory circuit Prakitchai Chotewutmontria and Alice Barkana,1 aInstitute of Molecular Biology, University of Oregon, Eugene, OR 97403 Edited by Krishna K. Niyogi, University of California, Berkeley, CA, and approved July 22, 2020 (received for review April 26, 2020) The D1 reaction center protein of photosystem II (PSII) is subject to mRNA to provide D1 for PSII repair remain obscure (13, 14). light-induced damage. Degradation of damaged D1 and its re- The consensus view in recent years has been that psbA transla- placement by nascent D1 are at the heart of a PSII repair cycle, tion for PSII repair is regulated at the elongation step (7, 15–17), without which photosynthesis is inhibited. In mature plant chloro- a view that arises primarily from experiments with the green alga plasts, light stimulates the recruitment of ribosomes specifically to Chlamydomonas reinhardtii (Chlamydomonas) (18). However, we psbA mRNA to provide nascent D1 for PSII repair and also triggers showed recently that regulated translation initiation makes a a global increase in translation elongation rate. The light-induced large contribution in plants (19). These experiments used ribo- signals that initiate these responses are unclear. We present action some profiling (ribo-seq) to monitor ribosome occupancy on spectrum and genetic data indicating that the light-induced re- cruitment of ribosomes to psbA mRNA is triggered by D1 photo- chloroplast open reading frames (ORFs) in maize and Arabi- damage, whereas the global stimulation of translation elongation dopsis upon shifting seedlings harboring mature chloroplasts is triggered by photosynthetic electron transport. -

ATP-Citrate Lyase Has an Essential Role in Cytosolic Acetyl-Coa Production in Arabidopsis Beth Leann Fatland Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2002 ATP-citrate lyase has an essential role in cytosolic acetyl-CoA production in Arabidopsis Beth LeAnn Fatland Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Molecular Biology Commons, and the Plant Sciences Commons Recommended Citation Fatland, Beth LeAnn, "ATP-citrate lyase has an essential role in cytosolic acetyl-CoA production in Arabidopsis " (2002). Retrospective Theses and Dissertations. 1218. https://lib.dr.iastate.edu/rtd/1218 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. ATP-citrate lyase has an essential role in cytosolic acetyl-CoA production in Arabidopsis by Beth LeAnn Fatland A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Plant Physiology Program of Study Committee: Eve Syrkin Wurtele (Major Professor) James Colbert Harry Homer Basil Nikolau Martin Spalding Iowa State University Ames, Iowa 2002 UMI Number: 3158393 INFORMATION TO USERS The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleed-through, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. -

Cell-Type-Specific Cytokinin Distribution Within The
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Spiral - Imperial College Digital Repository The Plant Cell, Vol. 27: 1955–1967, July 2015, www.plantcell.org ã 2015 American Society of Plant Biologists. All rights reserved. Cell-Type-Specific Cytokinin Distribution within the Arabidopsis Primary Root ApexOPEN Ioanna Antoniadi,a,b,1 Lenka Placková, c,1 Biljana Simonovik,a Karel Dolezal, c Colin Turnbull,b Karin Ljung,a,2 and Ondrej Novákc,2,3 a Umeå Plant Science Centre, Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences, SE-901 83 Umeå, Sweden b Department of Life Sciences, Imperial College London, London SW7 2AZ, United Kingdom c Laboratory of Growth Regulators and Department of Chemical Biology and Genetics, Centre of the Region Haná for Biotechnological and Agricultural Research, Institute of Experimental Botany AS CR and Faculty of Science of Palacký University, Slechtitelu˚ 27, CZ-78371 Olomouc, Czech Republic ORCID IDs: 0000-0001-9053-2788 (I.A.); 0000-0003-2537-4933 (L.P.); 0000-0002-0929-0791 (B.S.); 0000-0001-6635-1418 (C.T.); 0000-0003-2901-189X (K.L.); 0000-0003-3452-0154 (O.N.) Cytokinins (CKs) play a crucial role in many physiological and developmental processes at the levels of individual plant components (cells, tissues, and organs) and by coordinating activities across these parts. High-resolution measurements of intracellular CKs in different plant tissues can therefore provide insights into their metabolism and mode of action. Here, we applied fluorescence-activated cell sorting of green fluorescent protein (GFP)-marked cell types, combined with solid-phase microextraction and an ultra-high-sensitivity mass spectrometry (MS) method for analysis of CK biosynthesis and homeostasis at cellular resolution. -

Substrate Channel Flexibility in Pseudomonas Aeruginosa Murb Accommodates Two Distinct Substrates
Substrate Channel Flexibility in Pseudomonas aeruginosa MurB Accommodates Two Distinct Substrates Ming Wei Chen1,2, Bernhard Lohkamp1, Robert Schnell1, Julien Lescar2, Gunter Schneider1* 1 Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden, 2 School of Biological Sciences, Nanyang Technological University, Singapore, Singapore Abstract Biosynthesis of UDP-N-acetylmuramic acid in bacteria is a committed step towards peptidoglycan production. In an NADPH- and FAD-dependent reaction, the UDP-N-acetylglucosamine-enolpyruvate reductase (MurB) reduces UDP-N-acetylgluco- samine-enolpyruvate to UDP-N-acetylmuramic acid. We determined the three-dimensional structures of the ternary complex of Pseudomonas aeruginosa MurB with FAD and NADP+ in two crystal forms to resolutions of 2.2 and 2.1 A˚, respectively, to investigate the structural basis of the first half-reaction, hydride transfer from NADPH to FAD. The nicotinamide ring of NADP+ stacks against the si face of the isoalloxazine ring of FAD, suggesting an unusual mode of hydride transfer to flavin. Comparison with the structure of the Escherichia coli MurB complex with UDP-N- acetylglucosamine-enolpyruvate shows that both substrates share the binding site located between two lobes of the substrate-binding domain III, consistent with a ping pong mechanism with sequential substrate binding. The nicotinamide and the enolpyruvyl moieties are strikingly well-aligned upon superimposition, both positioned for hydride transfer to and from FAD. However, flexibility of the substrate channel allows the non-reactive parts of the two substrates to bind in different conformations. A potassium ion in the active site may assist in substrate orientation and binding. These structural models should help in structure-aided drug design against MurB, which is essential for cell wall biogenesis and hence bacterial survival. -

Negative Regulation of Diacylglycerol Kinase &Theta
Cell Death and Differentiation (2010) 17, 1059–1068 & 2010 Macmillan Publishers Limited All rights reserved 1350-9047/10 $32.00 www.nature.com/cdd Negative regulation of diacylglycerol kinase h mediates adenosine-dependent hepatocyte preconditioning G Baldanzi1,5, E Alchera2,5, C Imarisio2, M Gaggianesi1, C Dal Ponte2, M Nitti3, C Domenicotti3, WJ van Blitterswijk4, E Albano2, A Graziani1,5 and R Carini*,2,5 In liver ischemic preconditioning (IP), stimulation of adenosine A2a receptors (A2aR) prevents ischemia/reperfusion injury by promoting diacylglycerol-mediated activation of protein kinase C (PKC). By concerting diacylglycerol to phosphatidic acid, diacylglycerol kinases (DGKs) act as terminator of diacylglycerol signalling. This study investigates the role of DGK in the development of hepatocyte IP. DGK activity and cell viability were evaluated in isolated rat hepatocytes preconditioned by 10 min hypoxia followed by 10 min re-oxygenation or by the treatment with the A2aR agonist, CGS21680, and subsequently exposed to prolonged hypoxia. We observed that after IP or A2aR activation, a decrease in DGK activity was associated with the onset of hepatocyte tolerance to hypoxia. CGS21680-induced stimulation of A2aR specifically inhibited DGK isoform h by activating RhoA–GTPase. Consistently, both siRNA-mediated downregulation of DGK h and hepatocyte pretreatment with the DGK inhibitor R59949 induced cell tolerance to hypoxia. The pharmacological inhibition of DGK was associated with the diacylglycerol- dependent activation of PKC d and e and of their downstream target p38 MAPK. In conclusion, we unveil a novel signalling pathway contributing to the onset of hepatocyte preconditioning, which through RhoA–GTPase, couples A2aR to the downregulation of DGK. -

Properties of Chlorophyllase from Capsicum Annuum L. Fruits
Properties of Chlorophyllase from Capsicum annuum L. Fruits Dámaso Hornero-Méndez and Marí a Isabel Mínguez-Mosquera* Departamento de Biotecnologia de Alimentos, Instituto de la Grasa (CSIC), Av. Padre Garcia Tejero, 4, 41012-Sevilla, SPAIN. Fax: +34-954691262. E-mail: [email protected] * Author for correspondence and reprint requests Z. Naturforsch. 56c, 1015-1021 (2001); received June 27/August 6 , 2001 Chlorophyll, Chlorophyllase, Capsicum annuum The in vitro properties of semi-purified chlorophyllase (chlorophyll-chlorophyllido hy drolase, EC 3.1.1.14) from Capsicum annuum fruits have been studied. The enzyme showed an optimum of activity at pH 8.5 and 50 °C. Substrate specificity was studied for chlorophyll (Chi) a, Chi b, pheophytin (Phe) a and Phe b, with K m values of 10.70, 4.04, 2.67 and 6.37 ^im respectively. Substrate inhibition was found for Phe b at concentrations higher than 5 ^m. Chlorophyllase action on Chi a ’ and Chi b' was also studied but no hydrolysis was observed, suggesting that the mechanism of action depends on the configuration at C-132 in the chloro phyll molecule, with the enzyme acting only on compounds with R132 stereochemistry. The effect of various metals (Mg2+, Hg2+, Cu2+, Zn2+, Co , Fe2+ and Fe3+) was also investigated, and a general inhibitory effect was found, this being more marked for Hg2+ and Fe2+. Func tional groups such as -SH and -S-S- seemed to participate in the formation of the enzyme- substrate complex. Chelating ion and the carbonyl group at C3 appeared to be important in substrate recognition by the enzyme. -

Structure of Pigment Metabolic Pathways and Their Contributions to White Tepal Color Formation of Chinese Narcissus Tazetta Var
International Journal of Molecular Sciences Article Structure of Pigment Metabolic Pathways and Their Contributions to White Tepal Color Formation of Chinese Narcissus tazetta var. chinensis cv Jinzhanyintai Yujun Ren † ID , Jingwen Yang †, Bingguo Lu, Yaping Jiang, Haiyang Chen, Yuwei Hong, Binghua Wu and Ying Miao * Center for Molecular Cell and Systems Biology, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou 350002, China; [email protected] (Y.R.); [email protected] (J.Y.); [email protected] (B.L.); [email protected] (Y.J.); [email protected] (H.C.); [email protected] (Y.H.); [email protected] (B.W.) * Correspondence: [email protected]; Tel.:/Fax: +86-591-8639-2987 † These authors contributed equally to this work. Received: 7 August 2017; Accepted: 4 September 2017; Published: 8 September 2017 Abstract: Chinese narcissus (Narcissus tazetta var. chinensis) is one of the ten traditional flowers in China and a famous bulb flower in the world flower market. However, only white color tepals are formed in mature flowers of the cultivated varieties, which constrains their applicable occasions. Unfortunately, for lack of genome information of narcissus species, the explanation of tepal color formation of Chinese narcissus is still not clear. Concerning no genome information, the application of transcriptome profile to dissect biological phenomena in plants was reported to be effective. As known, pigments are metabolites of related metabolic pathways, which dominantly decide flower color. In this study, transcriptome profile and pigment metabolite analysis methods were used in the most widely cultivated Chinese narcissus “Jinzhanyintai” to discover the structure of pigment metabolic pathways and their contributions to white tepal color formation during flower development and pigmentation processes. -

Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma Divulgatum
microorganisms Article Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma divulgatum Rafael Bargiela 1 , Karin Lanthaler 1,2, Colin M. Potter 1,2 , Manuel Ferrer 3 , Alexander F. Yakunin 1,2, Bela Paizs 1,2, Peter N. Golyshin 1,2 and Olga V. Golyshina 1,2,* 1 School of Natural Sciences, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK; [email protected] (R.B.); [email protected] (K.L.); [email protected] (C.M.P.); [email protected] (A.F.Y.); [email protected] (B.P.); [email protected] (P.N.G.) 2 Centre for Environmental Biotechnology, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK 3 Systems Biotechnology Group, Department of Applied Biocatalysis, CSIC—Institute of Catalysis, Marie Curie 2, 28049 Madrid, Spain; [email protected] * Correspondence: [email protected]; Tel.: +44-1248-388607; Fax: +44-1248-382569 Received: 27 April 2020; Accepted: 15 May 2020; Published: 19 May 2020 Abstract: The archaeon Cuniculiplasma divulgatum is ubiquitous in acidic environments with low-to-moderate temperatures. However, molecular mechanisms underlying its ability to thrive at lower temperatures remain unexplored. Using mass spectrometry (MS)-based proteomics, we analysed the effect of short-term (3 h) exposure to cold. The C. divulgatum genome encodes 2016 protein-coding genes, from which 819 proteins were identified in the cells grown under optimal conditions. In line with the peptidolytic lifestyle of C. divulgatum, its intracellular proteome revealed the abundance of proteases, ABC transporters and cytochrome C oxidase. From 747 quantifiable polypeptides, the levels of 582 proteins showed no change after the cold shock, whereas 104 proteins were upregulated suggesting that they might be contributing to cold adaptation. -

Targeting Glioblastoma Stem Cells Through Disruption of the Circadian Clock
Published OnlineFirst August 27, 2019; DOI: 10.1158/2159-8290.CD-19-0215 RESEARCH ARTICLE Targeting Glioblastoma Stem Cells through Disruption of the Circadian Clock Zhen Dong1, Guoxin Zhang1, Meng Qu2, Ryan C. Gimple1,3, Qiulian Wu1, Zhixin Qiu1, Briana C. Prager1,3, Xiuxing Wang1, Leo J.Y. Kim1,3, Andrew R. Morton3, Deobrat Dixit1, Wenchao Zhou4, Haidong Huang4, Bin Li5, Zhe Zhu1, Shideng Bao4, Stephen C. Mack6, Lukas Chavez7, Steve A. Kay2, and Jeremy N. Rich1 Downloaded from cancerdiscovery.aacrjournals.org on September 24, 2021. © 2019 American Association for Cancer Research. Published OnlineFirst August 27, 2019; DOI: 10.1158/2159-8290.CD-19-0215 ABSTRACT Glioblastomas are highly lethal cancers, containing self-renewing glioblastoma stem cells (GSC). Here, we show that GSCs, differentiated glioblastoma cells (DGC), and nonmalignant brain cultures all displayed robust circadian rhythms, yet GSCs alone displayed exquisite dependence on core clock transcription factors, BMAL1 and CLOCK, for optimal cell growth. Downregulation of BMAL1 or CLOCK in GSCs induced cell-cycle arrest and apoptosis. Chromatin immu- noprecipitation revealed that BMAL1 preferentially bound metabolic genes and was associated with active chromatin regions in GSCs compared with neural stem cells. Targeting BMAL1 or CLOCK attenu- ated mitochondrial metabolic function and reduced expression of tricarboxylic acid cycle enzymes. Small-molecule agonists of two independent BMAL1–CLOCK negative regulators, the cryptochromes and REV-ERBs, downregulated stem cell factors and reduced GSC growth. Combination of cryp- tochrome and REV-ERB agonists induced synergistic antitumor effi cacy. Collectively, these fi ndings show that GSCs co-opt circadian regulators beyond canonical circadian circuitry to promote stemness maintenance and metabolism, offering novel therapeutic paradigms.