6ABIDA SECALE (Mollusca, Gastropoda, Pulmonata
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Compositional Variability of Pleistocene Land Snail Assemblages Preserved in a Cinder Cone
Compositional variability of Pleistocene land snail assemblages preserved in a cinder cone volcano from Tenerife, Canary Islands A thesis submitted to the graduate school of the University of Cincinnati in partial fulfillment of the requirements for the degree of Master of Science In the Department of Geology of the College of Arts and Sciences by Elizabeth M. Bullard B.S., Muskingum University, 2012 July 2016 Advisors: Dr. Yurena Yanes Dr. Arnold I. Miller Committee Member: Dr. Joshua Miller i Abstract Fossil assemblage faunal compositions may vary through space and time in response to climatic and/or taphonomic factors, but these relationships can be difficult to diagnose and disentangle. Here, we investigate how to disentangle climatic and taphonomic signals of a land- snail-rich volcanic scoria sequence to asses if it was influenced by taphonomic bias, climate change, or both, using a multifaceted approach, combining taphonomic, ecological, body-size, and stable-isotope data. Fossil assemblages were sampled from two beds (Units A and B) in a Pleistocene cinder cone volcano of southern Tenerife (Canary Islands), dated to the glacial interval MIS 8 (~299-302 ka). The two units differed in taphonomy, species composition, and abundance distributions. The upper unit, B (6 species), showed higher snail diversity and shell concentration and lower taphonomic alteration than the lower unit, A (3 species). Furthermore, larger bodied species (length>10mm) dominated Unit A and were better preserved than smaller species (length<10mm), whereas smaller individuals were more abundant (and better preserved) at Unit B. These differences were likely impacted by physical differences in the matrices surrounding the fossils. -

Molluscs of the Dürrenstein Wilderness Area
Molluscs of the Dürrenstein Wilderness Area S a b i n e F ISCHER & M i c h a e l D UDA Abstract: Research in the Dürrenstein Wilderness Area (DWA) in the southwest of Lower Austria is mainly concerned with the inventory of flora, fauna and habitats, interdisciplinary monitoring and studies on ecological disturbances and process dynamics. During a four-year qualitative study of non-marine molluscs, 96 sites within the DWA and nearby nature reserves were sampled in cooperation with the “Alpine Land Snails Working Group” located at the Natural History Museum of Vienna. Altogether, 84 taxa were recorded (72 land snails, 12 water snails and mussels) including four endemics and seven species listed in the Austrian Red List of Molluscs. A reference collection (empty shells) of molluscs, which is stored at the DWA administration, was created. This project was the first systematic survey of mollusc fauna in the DWA. Further sampling might provide additional information in the future, particularly for Hydrobiidae in springs and caves, where detailed analyses (e.g. anatomical and genetic) are needed. Key words: Wilderness Dürrenstein, Primeval forest, Benign neglect, Non-intervention management, Mollusca, Snails, Alpine endemics. Introduction manifold species living in the wilderness area – many of them “refugees”, whose natural habitats have almost In concordance with the IUCN guidelines, research is disappeared in today’s over-cultivated landscape. mandatory for category I wilderness areas. However, it may not disturb the natural habitats and communities of the nature reserve. Research in the Dürrenstein The Dürrenstein Wilderness Area Wilderness Area (DWA) focuses on providing invento- (DWA) ries of flora and fauna, on interdisciplinary monitoring The Dürrenstein Wilderness Area (DWA) was as well as on ecological disturbances and process dynamics. -

The Field Museum 2002 Annual Report to the Board of Trustees Academic Affairs
THE FIELD MUSEUM 2002 ANNUAL REPORT TO THE BOARD OF TRUSTEES ACADEMIC AFFAIRS Office of Academic Affairs, The Field Museum 1400 South Lake Shore Drive Chicago, IL 60605-2496 USA Phone (312) 665-7811 Fax (312) 665-7806 WWW address: http://www.fieldmuseum.org - This Report Printed on Recycled Paper - -1- Revised May 2003 -2- CONTENTS 2002 Annual Report....................................................................................................................................................3 Collections and Research Committee.....................................................................................................................12 Academic Affairs Staff List......................................................................................................................................13 Publications, 2002 .....................................................................................................................................................19 Active Grants, 2002...................................................................................................................................................38 Conferences, Symposia, Workshops and Invited Lectures, 2002 .......................................................................46 Museum and Public Service, 2002 ..........................................................................................................................55 Fieldwork and Research Travel, 2002 ....................................................................................................................65 -

Amaia Caro Aramendia
The genus Pyrenaearia (Gastropoda, Helicoidea): Molecular and Morphological Systematics, Biogeography and Population Dynamics Pyrenaearia generoa (Gastropoda, Helicoidea): Sistematika Molekularra eta Morfologikoa, Biogeografia eta Populazio Dinamika PhD thesis Vitoria-Gasteiz, 2019 Amaia Caro Aramendia The genus Pyrenaearia (Gastropoda, Helicoidea): Molecular and Morphological Systematics, Biogeography and Population Dynamics Pyrenaearia generoa (Gastropoda, Helicoidea): Sistematika Molekularra eta Morfologikoa, Biogeografia eta Populazio Dinamika A thesis submitted by Amaia Caro Aramendia for the degree of Doctor of Philosophy, under the supervision of Dr. Benjamín Juan Gómez-Moliner and Dr. María José Madeira University of the Basque Country, Vitoria-Gasteiz, 2019 Zoologia eta Animalia Biologia Zelulen Saila Dpto. Zoología y Biología Celular Animal (cc)2019 AMAIA CARO ARAMENDIA (cc by-nc-nd 4.0) Astiro igo, barraskilotxo Fuji mendia da hau! Kobayashi Issa-ren haikua To the little things that run the world Esker onak Acknowledgements Tesi bat ez da pertsona bakar batena, bidean zehar laguntzen duten pertsona guztiei esker sortutako lana da eta, beraz, lehen orriek haien laguntza eskertzeko izan behar dute: En primer lugar me gustaría agradecer a mis directores, Benjamín Gómez-Moliner y María José Madeira. A Benjamín, por darme la oportunidad de entrar en el grupo de investigación y confiar en que podría realizar esta tesis. Gracias por compartir tus extensos conocimientos y por descubrirme el mundo de la malacología, que sin duda no habría encontrado por mi cuenta y ha resultado de lo más interesante. A Marijo, porque desde el principio y hasta el final has estado siempre ahí para guiarme, animarme y para ayudarme en todo lo que hiciese falta pero, sobre todo, por mostrarme que es posible compaginar este trabajo con una vida fuera de él. -

293 – Les Noms Scientifiques Français Des Mollusques Continentaux De
LES NOMS SCIENTIFIQUES FRANÇAIS DES MOLLUSQUES CONTINENTAUX DE FRANCE : processus d’ÉtABLISSEMENT d’une LISTE DE RÉFÉRENCE Benoît FONTAINE 1, Jean-Michel BICHAIN 2, Xavier CUCHERAT 3, Olivier GAR G OMINY 4 & Vincent PRIÉ 5 SUMMARY. — French scientific names of continental molluscs of France: process for establishing a list of reference. — In the biodiversity crisis context and with the increasing general awareness on this issue, conservation of small and poorly-known species is hampered by the fact they only have latine names. In order to communicate for biodiversity conservation, having French names is an advantage which is lacking in terrestrial and freshwater molluscs from France. To remedy this problem, we propose a list of French scientific names for this group, i.e. all species and subspecies known from France. We have listed existing names in legal documents, in usage and in the 18th and 19th centuries scientific literature. The resulting list being incomplete, we had to create new French names, following a series of recommendations adapted from similar works dealing with other taxonomic groups. We conclude by dealing with the issue of the legitimacy and validity of such names. The list of French scientific names is given as an appendix and is downloadable from internet. RÉSUMÉ. — Dans le contexte de la crise de la biodiversité et de la prise de conscience par le grand public des enjeux environnementaux, la conservation des espèces petites et méconnues est handicapée par le fait que ces espèces ne peuvent être désignées que par leur nom latin. Dans une optique de communication pour la préservation de la biodiversité, disposer de nom français est un atout qui fait défaut pour les mol- lusques terrestres et d’eau douce de France. -

(Approx) Mixed Micro Shells (22G Bags) Philippines € 10,00 £8,64 $11,69 Each 22G Bag Provides Hours of Fun; Some Interesting Foraminifera Also Included
Special Price £ US$ Family Genus, species Country Quality Size Remarks w/o Photo Date added Category characteristic (€) (approx) (approx) Mixed micro shells (22g bags) Philippines € 10,00 £8,64 $11,69 Each 22g bag provides hours of fun; some interesting Foraminifera also included. 17/06/21 Mixed micro shells Ischnochitonidae Callistochiton pulchrior Panama F+++ 89mm € 1,80 £1,55 $2,10 21/12/16 Polyplacophora Ischnochitonidae Chaetopleura lurida Panama F+++ 2022mm € 3,00 £2,59 $3,51 Hairy girdles, beautifully preserved. Web 24/12/16 Polyplacophora Ischnochitonidae Ischnochiton textilis South Africa F+++ 30mm+ € 4,00 £3,45 $4,68 30/04/21 Polyplacophora Ischnochitonidae Ischnochiton textilis South Africa F+++ 27.9mm € 2,80 £2,42 $3,27 30/04/21 Polyplacophora Ischnochitonidae Stenoplax limaciformis Panama F+++ 16mm+ € 6,50 £5,61 $7,60 Uncommon. 24/12/16 Polyplacophora Chitonidae Acanthopleura gemmata Philippines F+++ 25mm+ € 2,50 £2,16 $2,92 Hairy margins, beautifully preserved. 04/08/17 Polyplacophora Chitonidae Acanthopleura gemmata Australia F+++ 25mm+ € 2,60 £2,25 $3,04 02/06/18 Polyplacophora Chitonidae Acanthopleura granulata Panama F+++ 41mm+ € 4,00 £3,45 $4,68 West Indian 'fuzzy' chiton. Web 24/12/16 Polyplacophora Chitonidae Acanthopleura granulata Panama F+++ 32mm+ € 3,00 £2,59 $3,51 West Indian 'fuzzy' chiton. 24/12/16 Polyplacophora Chitonidae Chiton tuberculatus Panama F+++ 44mm+ € 5,00 £4,32 $5,85 Caribbean. 24/12/16 Polyplacophora Chitonidae Chiton tuberculatus Panama F++ 35mm € 2,50 £2,16 $2,92 Caribbean. 24/12/16 Polyplacophora Chitonidae Chiton tuberculatus Panama F+++ 29mm+ € 3,00 £2,59 $3,51 Caribbean. -

Draft Carpathian Red List of Forest Habitats
CARPATHIAN RED LIST OF FOREST HABITATS AND SPECIES CARPATHIAN LIST OF INVASIVE ALIEN SPECIES (DRAFT) PUBLISHED BY THE STATE NATURE CONSERVANCY OF THE SLOVAK REPUBLIC 2014 zzbornik_cervenebornik_cervene zzoznamy.inddoznamy.indd 1 227.8.20147.8.2014 222:36:052:36:05 © Štátna ochrana prírody Slovenskej republiky, 2014 Editor: Ján Kadlečík Available from: Štátna ochrana prírody SR Tajovského 28B 974 01 Banská Bystrica Slovakia ISBN 978-80-89310-81-4 Program švajčiarsko-slovenskej spolupráce Swiss-Slovak Cooperation Programme Slovenská republika This publication was elaborated within BioREGIO Carpathians project supported by South East Europe Programme and was fi nanced by a Swiss-Slovak project supported by the Swiss Contribution to the enlarged European Union and Carpathian Wetlands Initiative. zzbornik_cervenebornik_cervene zzoznamy.inddoznamy.indd 2 115.9.20145.9.2014 223:10:123:10:12 Table of contents Draft Red Lists of Threatened Carpathian Habitats and Species and Carpathian List of Invasive Alien Species . 5 Draft Carpathian Red List of Forest Habitats . 20 Red List of Vascular Plants of the Carpathians . 44 Draft Carpathian Red List of Molluscs (Mollusca) . 106 Red List of Spiders (Araneae) of the Carpathian Mts. 118 Draft Red List of Dragonfl ies (Odonata) of the Carpathians . 172 Red List of Grasshoppers, Bush-crickets and Crickets (Orthoptera) of the Carpathian Mountains . 186 Draft Red List of Butterfl ies (Lepidoptera: Papilionoidea) of the Carpathian Mts. 200 Draft Carpathian Red List of Fish and Lamprey Species . 203 Draft Carpathian Red List of Threatened Amphibians (Lissamphibia) . 209 Draft Carpathian Red List of Threatened Reptiles (Reptilia) . 214 Draft Carpathian Red List of Birds (Aves). 217 Draft Carpathian Red List of Threatened Mammals (Mammalia) . -

Does Postfire Management Affect the Recovery of Mediterranean
Forest Ecology and Management 261 (2011) 611–619 Contents lists available at ScienceDirect Forest Ecology and Management journal homepage: www.elsevier.com/locate/foreco Does postfire management affect the recovery of Mediterranean communities? The case study of terrestrial gastropods Vicenc¸ Bros a, Gregorio Moreno-Rueda b, Xavier Santos c,∗ a Parc Natural de Sant Llorenc¸ del Munt i l’Obac, Oficina Tècnica de Parcs Naturals, Diputació de Barcelona, c/Urgell 187, Edif. Rellotge 3a, E-08036 Barcelona, Spain b Estación Experimental de Zonas Áridas (CSIC), La Ca˜nada de San Urbano, Ctra. Sacramento s/n, E-04120 Almería, Spain c Departament de Biologia Animal, Universitat de Barcelona, Av. Diagonal 645, E-08028 Barcelona, Spain article info abstract Article history: In fire-prone regions, understanding the response of species to fire is a major goal in order to predict Received 8 September 2010 the effects on biodiversity. Furthermore, postfire management can also model this response through the Received in revised form manipulation of environmental characteristics of the burnt habitat. We have examined the taxonomic 10 November 2010 and functional response to fire and postfire management of a Mediterranean snail community affected Accepted 13 November 2010 by a summer fire in 2003. After the fire, the area was logged, leaving wood debris on the ground, and Available online 16 December 2010 three alternative practices were implemented in several plots within the burnt area: subsoiling, removal of trunks having branches, total removal of trunks and branches, as well as one area not logged. Our Keywords: Mollusca results indicated that fire exerted a major impact on the snail community, strongly reducing diversity Terrestrial snails and species richness, particularly for forest species living in the humus and having European distribution Postfire management ranges. -

Biodiversity of the Hypersaline Urmia Lake National Park (NW Iran)
Diversity 2014, 6, 102-132; doi:10.3390/d6010102 OPEN ACCESS diversity ISSN 1424-2818 www.mdpi.com/journal/diversity Review Biodiversity of the Hypersaline Urmia Lake National Park (NW Iran) Alireza Asem 1,†,*, Amin Eimanifar 2,†,*, Morteza Djamali 3, Patricio De los Rios 4 and Michael Wink 2 1 Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao 266003, China 2 Institute of Pharmacy and Molecular Biotechnology (IPMB), Heidelberg University, Im Neuenheimer Feld 364, Heidelberg D-69120, Germany; E-Mail: [email protected] 3 Institut Méditerranéen d’Ecologie et de Paléoécologie UMR 6116 du CNRS-Europôle Méditerranéen de l’Arbois-Pavillon Villemin-BP 80, Aix-en-Provence Cedex 04 13545, France; E-Mail: [email protected] 4 Environmental Sciences School, Natural Resources Faculty, Catholic University of Temuco, Casilla 15-D, Temuco 4780000, Chile; E-Mail: [email protected] † These authors contributed equally to this work. * Authors to whom correspondence should be addressed; E-Mails: [email protected] (A.A.); [email protected] (A.E.); Tel.: +86-150-6624-4312 (A.A.); Fax: +86-532-8203-2216 (A.A.); Tel.: +49-6221-544-880 (A.E.); Fax: +49-6221-544-884 (A.E.). Received: 3 December 2013; in revised form: 13 January 2014 / Accepted: 27 January 2014 / Published: 10 February 2014 Abstract: Urmia Lake, with a surface area between 4000 to 6000 km2, is a hypersaline lake located in northwest Iran. It is the saltiest large lake in the world that supports life. Urmia Lake National Park is the home of an almost endemic crustacean species known as the brine shrimp, Artemia urmiana. -
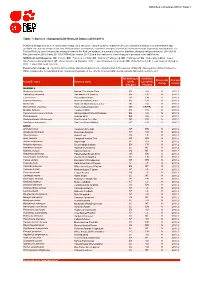
Table 7: Species Changing IUCN Red List Status (2010-2011)
IUCN Red List version 2011.2: Table 7 Table 7: Species changing IUCN Red List Status (2010-2011) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2010 (IUCN Red List version 2010.4) and 2011 (IUCN Red List version 2011.2) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered, EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.) IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2010) List (2011) change version Category Category MAMMALS Bradypus torquatus Maned Three-toed Sloth EN VU N 2011.1 Callicebus oenanthe San Martin Titi Monkey EN CR N 2011.1 Equus ferus Przewalski's Horse CR EN G 2011.2 -

Fauna of New Zealand Website Copy 2010, Fnz.Landcareresearch.Co.Nz
aua o ew eaa Ko te Aiaga eeke o Aoeaoa IEEAE SYSEMAICS AISOY GOU EESEAIES O ACAE ESEAC ema acae eseac ico Agicuue & Sciece Cee P O o 9 ico ew eaa K Cosy a M-C aiièe acae eseac Mou Ae eseac Cee iae ag 917 Aucka ew eaa EESEAIE O UIESIIES M Emeso eame o Eomoogy & Aima Ecoogy PO o ico Uiesiy ew eaa EESEAIE O MUSEUMS M ama aua Eiome eame Museum o ew eaa e aa ogaewa O o 7 Weigo ew eaa EESEAIE O OESEAS ISIUIOS awece CSIO iisio o Eomoogy GO o 17 Caea Ciy AC 1 Ausaia SEIES EIO AUA O EW EAA M C ua (ecease ue 199 acae eseac Mou Ae eseac Cee iae ag 917 Aucka ew eaa Fauna of New Zealand Ko te Aitanga Pepeke o Aotearoa Number / Nama 38 Naturalised terrestrial Stylommatophora (Mousca Gasooa Gay M ake acae eseac iae ag 317 amio ew eaa 4 Maaaki Whenua Ρ Ε S S ico Caeuy ew eaa 1999 Coyig © acae eseac ew eaa 1999 o a o is wok coee y coyig may e eouce o coie i ay om o y ay meas (gaic eecoic o mecaica icuig oocoyig ecoig aig iomaio eiea sysems o oewise wiou e wie emissio o e uise Caaoguig i uicaio AKE G Μ (Gay Micae 195— auase eesia Syommaooa (Mousca Gasooa / G Μ ake — ico Caeuy Maaaki Weua ess 1999 (aua o ew eaa ISS 111-533 ; o 3 IS -7-93-5 I ie 11 Seies UC 593(931 eae o uIicaio y e seies eio (a comee y eo Cosy usig comue-ase e ocessig ayou scaig a iig a acae eseac M Ae eseac Cee iae ag 917 Aucka ew eaa Māoi summay e y aco uaau Cosuas Weigo uise y Maaaki Weua ess acae eseac O o ico Caeuy Wesie //wwwmwessco/ ie y G i Weigo o coe eoceas eicuaum (ue a eigo oaa (owe (IIusao G M ake oucio o e coou Iaes was ue y e ew eaIa oey oa ue oeies eseac -

Guidelines for the Capture and Management of Digital Zoological Names Information Francisco W
Guidelines for the Capture and Management of Digital Zoological Names Information Francisco W. Welter-Schultes Version 1.1 March 2013 Suggested citation: Welter-Schultes, F.W. (2012). Guidelines for the capture and management of digital zoological names information. Version 1.1 released on March 2013. Copenhagen: Global Biodiversity Information Facility, 126 pp, ISBN: 87-92020-44-5, accessible online at http://www.gbif.org/orc/?doc_id=2784. ISBN: 87-92020-44-5 (10 digits), 978-87-92020-44-4 (13 digits). Persistent URI: http://www.gbif.org/orc/?doc_id=2784. Language: English. Copyright © F. W. Welter-Schultes & Global Biodiversity Information Facility, 2012. Disclaimer: The information, ideas, and opinions presented in this publication are those of the author and do not represent those of GBIF. License: This document is licensed under Creative Commons Attribution 3.0. Document Control: Version Description Date of release Author(s) 0.1 First complete draft. January 2012 F. W. Welter- Schultes 0.2 Document re-structured to improve February 2012 F. W. Welter- usability. Available for public Schultes & A. review. González-Talaván 1.0 First public version of the June 2012 F. W. Welter- document. Schultes 1.1 Minor editions March 2013 F. W. Welter- Schultes Cover Credit: GBIF Secretariat, 2012. Image by Levi Szekeres (Romania), obtained by stock.xchng (http://www.sxc.hu/photo/1389360). March 2013 ii Guidelines for the management of digital zoological names information Version 1.1 Table of Contents How to use this book ......................................................................... 1 SECTION I 1. Introduction ................................................................................ 2 1.1. Identifiers and the role of Linnean names ......................................... 2 1.1.1 Identifiers ..................................................................................