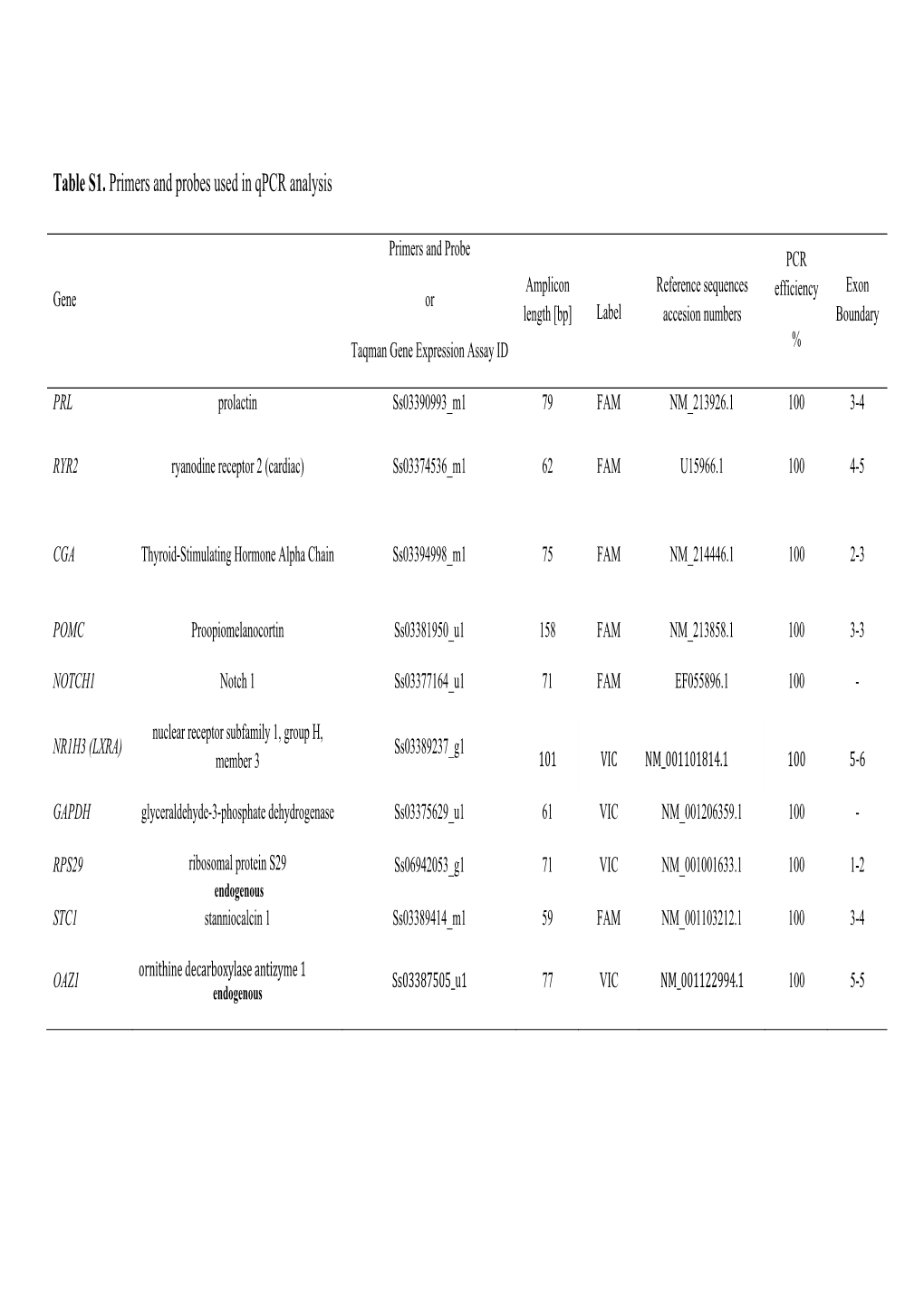
Table S1. Primers and Probes Used in Qpcr Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

3.2 Brain White Matter Oedema Due to Clc-2 Chloride Channel Deficiency
3.2 Brain white matter oedema due to ClC-2 chloride channel deficiency: an observational analytical study M Bugiani*, C Depienne*, C Dupuits, D Galanaud, V Touitou, N Postma, C van Berkel, E Polder, E Tollard, F Darios, A Brice, CE de Die-Smulders, JS Vles, A Vanderver, G Uziel, C Yalcinkaya, SG Frints, VM Kalscheuer, J Klooster, M Kamermans, TEM Abbink, NI Wolf, F Sedel** and MS van der Knaap** (*shared first authors; **shared last authors) Lancet Neurol 2013;12:659-668 Summary Background. Mutant mouse models suggest that the chloride channel ClC-2 has functions in ion and water homoeostasis, but this has not been confirmed in human beings. We aimed to define novel disorders characterised by distinct patterns of MRI abnormalities in patients with leukoencephalopathies of unknown origin, and to identify the genes mutated in these disorders. We were specifically interested in leukoencephalopathies characterised by white matter oedema, suggesting a defect in ion and water homoeostasis. Methods. In this observational analytical study, we recruited patients with leukoencephalopathies characterised by MRI signal abnormalities in the posterior limbs of the internal capsules, midbrain cerebral peduncles, and middle cerebellar peduncles from our databases of patients with leukoencephalopathies of unknown origin. We used exome sequencing to identify the gene involved. We screened the candidate gene in additional patients by Sanger sequencing and mRNA analysis, and investigated the functional effects of the mutations. We assessed the localisation of ClC-2 with immunohistochemistry and electron microscopy in post- mortem human brains of individuals without neurological disorders. Findings. Seven patients met our inclusion criteria, three with adult-onset disease and four with childhood-onset disease. -

The Mineralocorticoid Receptor Leads to Increased Expression of EGFR
www.nature.com/scientificreports OPEN The mineralocorticoid receptor leads to increased expression of EGFR and T‑type calcium channels that support HL‑1 cell hypertrophy Katharina Stroedecke1,2, Sandra Meinel1,2, Fritz Markwardt1, Udo Kloeckner1, Nicole Straetz1, Katja Quarch1, Barbara Schreier1, Michael Kopf1, Michael Gekle1 & Claudia Grossmann1* The EGF receptor (EGFR) has been extensively studied in tumor biology and recently a role in cardiovascular pathophysiology was suggested. The mineralocorticoid receptor (MR) is an important efector of the renin–angiotensin–aldosterone‑system and elicits pathophysiological efects in the cardiovascular system; however, the underlying molecular mechanisms are unclear. Our aim was to investigate the importance of EGFR for MR‑mediated cardiovascular pathophysiology because MR is known to induce EGFR expression. We identifed a SNP within the EGFR promoter that modulates MR‑induced EGFR expression. In RNA‑sequencing and qPCR experiments in heart tissue of EGFR KO and WT mice, changes in EGFR abundance led to diferential expression of cardiac ion channels, especially of the T‑type calcium channel CACNA1H. Accordingly, CACNA1H expression was increased in WT mice after in vivo MR activation by aldosterone but not in respective EGFR KO mice. Aldosterone‑ and EGF‑responsiveness of CACNA1H expression was confrmed in HL‑1 cells by Western blot and by measuring peak current density of T‑type calcium channels. Aldosterone‑induced CACNA1H protein expression could be abrogated by the EGFR inhibitor AG1478. Furthermore, inhibition of T‑type calcium channels with mibefradil or ML218 reduced diameter, volume and BNP levels in HL‑1 cells. In conclusion the MR regulates EGFR and CACNA1H expression, which has an efect on HL‑1 cell diameter, and the extent of this regulation seems to depend on the SNP‑216 (G/T) genotype. -

Structural Characterization of the Human Eukaryotic Initiation Factor 3 Protein Complex by Mass Spectrometry*□S
Supplemental Material can be found at: http://www.mcponline.org/cgi/content/full/M600399-MCP200 /DC1 Research Structural Characterization of the Human Eukaryotic Initiation Factor 3 Protein Complex by Mass Spectrometry*□S Eugen Damoc‡, Christopher S. Fraser§, Min Zhou¶, Hortense Videler¶, Greg L. Mayeurʈ, John W. B. Hersheyʈ, Jennifer A. Doudna§, Carol V. Robinson¶**, and Julie A. Leary‡ ‡‡ Protein synthesis in mammalian cells requires initiation The initiation phase of eukaryotic protein synthesis involves factor eIF3, an ϳ800-kDa protein complex that plays a formation of an 80 S ribosomal complex containing the initi- Downloaded from central role in binding of initiator methionyl-tRNA and ator methionyl-tRNAi bound to the initiation codon in the mRNA to the 40 S ribosomal subunit to form the 48 S mRNA. This is a multistep process promoted by proteins initiation complex. The eIF3 complex also prevents pre- called eukaryotic initiation factors (eIFs).1 Currently at least 12 mature association of the 40 and 60 S ribosomal subunits eIFs, composed of at least 29 distinct subunits, have been and interacts with other initiation factors involved in start identified (1). Mammalian eIF3, the largest initiation factor, is a codon selection. The molecular mechanisms by which multisubunit complex with an apparent molecular mass of www.mcponline.org eIF3 exerts these functions are poorly understood. Since ϳ800 kDa. This protein complex plays an essential role in its initial characterization in the 1970s, the exact size, translation by binding directly to the 40 S ribosomal subunit composition, and post-translational modifications of and promoting formation of the 43 S preinitiation complex ⅐ ⅐ mammalian eIF3 have not been rigorously determined. -

The Purification and Identification of Interactors to Elucidate Novel Connections in the HEK 293 Cell Line
The Purification and Identification of Interactors to Elucidate Novel Connections in the HEK 293 Cell Line Brett Hawley Biochemistry, Microbiology and Immunology Faculty of Medicine University of Ottawa © Brett Hawley, Ottawa, Canada, 2012 ABSTRACT The field of proteomics studies the structure and function of proteins in a large scale and high throughput manner. My work in the field of proteomics focuses on identifying interactions between proteins and discovering novel interactions. The identification of these interactions provides new information on metabolic and disease pathways and the working proteome of a cell. Cells are lysed and purified using antibody based affinity purification followed by digestion and identification using an HPLC coupled to a mass spectrometer. In my studies, I looked at the interaction networks of several AD related genes (Apolipoprotein E, Clusterin variant 1 and 2, Low-density lipoprotein receptor, Phosphatidylinositol binding clathrin assembly protein, Alpha- synuclein and Platelet-activating factor receptor) and an endosomal recycling pathway involved in cholesterol metabolism (Eps15 homology domain 1,2 and 4, Proprotein convertase subtilisin/kexin type 9 and Low-density lipoprotein receptor). Several novel and existing interactors were identified and these interactions were validated using co-immunopurification, which could be the basis for future research. ii ACKNOWLEDGEMENTS I would like to take this opportunity to thank my supervisor, Dr. Daniel Figeys, for his support and guidance throughout my studies in his lab. It was a great experience to work in his lab and I am very thankful I was given the chance to learn and work under him. I would also like to thank the members of my lab for all their assistance in learning new techniques and equipment in the lab. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Dependent Traits in Mice Models of Hyperthyroidism and Hypothyroidism
Phenotypical characterization of sex- dependent traits in mice models of hyperthyroidism and hypothyroidism Inaugural-Dissertation zur Erlangung des Doktorgrades Dr. rer. nat. der Fakultät für Biologie an der Universität Duisburg-Essen vorgelegt von Helena Rakov aus St. Petropawlowsk April 2017 Die der vorliegenden Arbeit zugrunde liegenden Experimente wurden am Universitätsklinikum Essen in der Klinik für Endokrinologie und Stoffwechselerkrankungen durchgeführt. 1. Gutachter: Prof. Dr. Dr. Dagmar Führer-Sakel 2. Gutachter: Prof. Dr. Elke Cario Vorsitzender des Prüfungsausschusses: Prof. Dr. Ruth Grümmer Tag der mündlichen Prüfung: 17.07.2017 Publications Publications Engels Kathrin*, Rakov Helena *, Zwanziger Denise, Moeller Lars C., Homuth Georg, Köhrle Josef, Brix Klaudia, Fuhrer Dagmar. Differences in mouse hepatic thyroid hormone transporter expression with age and hyperthyroidism. Eur Thyroid J 2015;4(suppl 1):81–86. DOI: 10.1159/000381020. *contributed equally Zwanziger Denise*, Rakov Helena*, Engels Kathrin, Moeller Lars C., Fuhrer Dagmar. Sex-dependent claudin-1 expression in liver of eu- and hypothyroid mice. Eur Thyroid J. 2015 Sep; 4(Suppl 1): 67–73. DOI: 10.1159/000431316. *contributed equally Engels Kathrin*, Rakov Helena*, Zwanziger Denise, Hoenes Georg Sebastian, Rehders Maren, Brix Klaudia, Koehrle Josef, Moeller Lars Christian, Fuhrer Dagmar. Efficacy of protocols for induction of chronic hyperthyroidism in male and female mice. Endocrine. 2016 Oct;54(1):47-54. DOI: 10.1007/s12020-016-1020-8. Rakov Helena*, Engels Kathrin*, Hönes Georg Sebastian, Strucksberg Karl-Heinz, Moeller Lars Christian, Köhrle Josef, Zwanziger Denise, Führer Dagmar. Sex-specific phenotypes of hyperthyroidism and hypothyroidism in mice. Biol Sex Differ. 2016 Aug 24;7(1):36. DOI: 10.1186/s13293-016-0089-3. -

Expression Profiling of Ion Channel Genes Predicts Clinical Outcome in Breast Cancer
UCSF UC San Francisco Previously Published Works Title Expression profiling of ion channel genes predicts clinical outcome in breast cancer Permalink https://escholarship.org/uc/item/1zq9j4nw Journal Molecular Cancer, 12(1) ISSN 1476-4598 Authors Ko, Jae-Hong Ko, Eun A Gu, Wanjun et al. Publication Date 2013-09-22 DOI http://dx.doi.org/10.1186/1476-4598-12-106 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Ko et al. Molecular Cancer 2013, 12:106 http://www.molecular-cancer.com/content/12/1/106 RESEARCH Open Access Expression profiling of ion channel genes predicts clinical outcome in breast cancer Jae-Hong Ko1, Eun A Ko2, Wanjun Gu3, Inja Lim1, Hyoweon Bang1* and Tong Zhou4,5* Abstract Background: Ion channels play a critical role in a wide variety of biological processes, including the development of human cancer. However, the overall impact of ion channels on tumorigenicity in breast cancer remains controversial. Methods: We conduct microarray meta-analysis on 280 ion channel genes. We identify candidate ion channels that are implicated in breast cancer based on gene expression profiling. We test the relationship between the expression of ion channel genes and p53 mutation status, ER status, and histological tumor grade in the discovery cohort. A molecular signature consisting of ion channel genes (IC30) is identified by Spearman’s rank correlation test conducted between tumor grade and gene expression. A risk scoring system is developed based on IC30. We test the prognostic power of IC30 in the discovery and seven validation cohorts by both Cox proportional hazard regression and log-rank test. -

Role of the Wnt/ B-Catenin Pathway in Liver Development and Zonation
University of Bath PHD Role of the Wnt/ -catenin Pathway in Liver Development and Zonation Shen Wen, Yeh Award date: 2012 Awarding institution: University of Bath Link to publication Alternative formats If you require this document in an alternative format, please contact: [email protected] General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 10. Oct. 2021 Role of the Wnt/ -catenin Pathway in Liver Development and Zonation YEH, SHENG-WEN A thesis submitted for the degree of Doctor of Philosophy University of Bath Department of Biology and Biochemistry September, 2012 COPYRIGHT Attention is drawn to the fact that copyright of this thesis rests with the author. A copy of this thesis has been supplied on condition that anyone who consults it is understood to recognise that its copyright rests with the author and that they must not copy it or use material from it except as permitted by law or with the consent of the author. -

Genes with 5' Terminal Oligopyrimidine Tracts Preferentially Escape Global Suppression of Translation by the SARS-Cov-2 NSP1 Protein
Downloaded from rnajournal.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Genes with 5′ terminal oligopyrimidine tracts preferentially escape global suppression of translation by the SARS-CoV-2 Nsp1 protein Shilpa Raoa, Ian Hoskinsa, Tori Tonna, P. Daniela Garciaa, Hakan Ozadama, Elif Sarinay Cenika, Can Cenika,1 a Department of Molecular Biosciences, University of Texas at Austin, Austin, TX 78712, USA 1Corresponding author: [email protected] Key words: SARS-CoV-2, Nsp1, MeTAFlow, translation, ribosome profiling, RNA-Seq, 5′ TOP, Ribo-Seq, gene expression 1 Downloaded from rnajournal.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract Viruses rely on the host translation machinery to synthesize their own proteins. Consequently, they have evolved varied mechanisms to co-opt host translation for their survival. SARS-CoV-2 relies on a non-structural protein, Nsp1, for shutting down host translation. However, it is currently unknown how viral proteins and host factors critical for viral replication can escape a global shutdown of host translation. Here, using a novel FACS-based assay called MeTAFlow, we report a dose-dependent reduction in both nascent protein synthesis and mRNA abundance in cells expressing Nsp1. We perform RNA-Seq and matched ribosome profiling experiments to identify gene-specific changes both at the mRNA expression and translation level. We discover that a functionally-coherent subset of human genes are preferentially translated in the context of Nsp1 expression. These genes include the translation machinery components, RNA binding proteins, and others important for viral pathogenicity. Importantly, we uncovered a remarkable enrichment of 5′ terminal oligo-pyrimidine (TOP) tracts among preferentially translated genes. -

Anti-Inflammatory Role of Curcumin in LPS Treated A549 Cells at Global Proteome Level and on Mycobacterial Infection
Anti-inflammatory Role of Curcumin in LPS Treated A549 cells at Global Proteome level and on Mycobacterial infection. Suchita Singh1,+, Rakesh Arya2,3,+, Rhishikesh R Bargaje1, Mrinal Kumar Das2,4, Subia Akram2, Hossain Md. Faruquee2,5, Rajendra Kumar Behera3, Ranjan Kumar Nanda2,*, Anurag Agrawal1 1Center of Excellence for Translational Research in Asthma and Lung Disease, CSIR- Institute of Genomics and Integrative Biology, New Delhi, 110025, India. 2Translational Health Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, 110067, India. 3School of Life Sciences, Sambalpur University, Jyoti Vihar, Sambalpur, Orissa, 768019, India. 4Department of Respiratory Sciences, #211, Maurice Shock Building, University of Leicester, LE1 9HN 5Department of Biotechnology and Genetic Engineering, Islamic University, Kushtia- 7003, Bangladesh. +Contributed equally for this work. S-1 70 G1 S 60 G2/M 50 40 30 % of cells 20 10 0 CURI LPSI LPSCUR Figure S1: Effect of curcumin and/or LPS treatment on A549 cell viability A549 cells were treated with curcumin (10 µM) and/or LPS or 1 µg/ml for the indicated times and after fixation were stained with propidium iodide and Annexin V-FITC. The DNA contents were determined by flow cytometry to calculate percentage of cells present in each phase of the cell cycle (G1, S and G2/M) using Flowing analysis software. S-2 Figure S2: Total proteins identified in all the three experiments and their distribution betwee curcumin and/or LPS treated conditions. The proteins showing differential expressions (log2 fold change≥2) in these experiments were presented in the venn diagram and certain number of proteins are common in all three experiments. -

Transcriptome-Wide Profiling of Cerebral Cavernous Malformations
www.nature.com/scientificreports OPEN Transcriptome-wide Profling of Cerebral Cavernous Malformations Patients Reveal Important Long noncoding RNA molecular signatures Santhilal Subhash 2,8, Norman Kalmbach3, Florian Wegner4, Susanne Petri4, Torsten Glomb5, Oliver Dittrich-Breiholz5, Caiquan Huang1, Kiran Kumar Bali6, Wolfram S. Kunz7, Amir Samii1, Helmut Bertalanfy1, Chandrasekhar Kanduri2* & Souvik Kar1,8* Cerebral cavernous malformations (CCMs) are low-fow vascular malformations in the brain associated with recurrent hemorrhage and seizures. The current treatment of CCMs relies solely on surgical intervention. Henceforth, alternative non-invasive therapies are urgently needed to help prevent subsequent hemorrhagic episodes. Long non-coding RNAs (lncRNAs) belong to the class of non-coding RNAs and are known to regulate gene transcription and involved in chromatin remodeling via various mechanism. Despite accumulating evidence demonstrating the role of lncRNAs in cerebrovascular disorders, their identifcation in CCMs pathology remains unknown. The objective of the current study was to identify lncRNAs associated with CCMs pathogenesis using patient cohorts having 10 CCM patients and 4 controls from brain. Executing next generation sequencing, we performed whole transcriptome sequencing (RNA-seq) analysis and identifed 1,967 lncRNAs and 4,928 protein coding genes (PCGs) to be diferentially expressed in CCMs patients. Among these, we selected top 6 diferentially expressed lncRNAs each having signifcant correlative expression with more than 100 diferentially expressed PCGs. The diferential expression status of the top lncRNAs, SMIM25 and LBX2-AS1 in CCMs was further confrmed by qRT-PCR analysis. Additionally, gene set enrichment analysis of correlated PCGs revealed critical pathways related to vascular signaling and important biological processes relevant to CCMs pathophysiology. -

Endothelial Loss of Fzd5 Stimulates PKC/Ets1-Mediated Transcription of Angpt2 and Flt1
Angiogenesis https://doi.org/10.1007/s10456-018-9625-6 ORIGINAL PAPER Endothelial loss of Fzd5 stimulates PKC/Ets1-mediated transcription of Angpt2 and Flt1 Maarten M. Brandt1 · Christian G. M. van Dijk2 · Ihsan Chrifi1 · Heleen M. Kool3 · Petra E. Bürgisser3 · Laura Louzao‑Martinez2 · Jiayi Pei2 · Robbert J. Rottier3 · Marianne C. Verhaar2 · Dirk J. Duncker1 · Caroline Cheng1,2 Received: 19 January 2018 / Accepted: 22 May 2018 © The Author(s) 2018 Abstract Aims Formation of a functional vascular system is essential and its formation is a highly regulated process initiated during embryogenesis, which continues to play important roles throughout life in both health and disease. In previous studies, Fzd5 was shown to be critically involved in this process and here we investigated the molecular mechanism by which endothelial loss of this receptor attenuates angiogenesis. Methods and results Using short interference RNA-mediated loss-of-function assays, the function and mechanism of sign- aling via Fzd5 was studied in human endothelial cells (ECs). Our findings indicate that Fzd5 signaling promotes neoves- sel formation in vitro in a collagen matrix-based 3D co-culture of primary vascular cells. Silencing of Fzd5 reduced EC proliferation, as a result of G 0/G1 cell cycle arrest, and decreased cell migration. Furthermore, Fzd5 knockdown resulted in enhanced expression of the factors Angpt2 and Flt1, which are mainly known for their destabilizing effects on the vasculature. In Fzd5-silenced ECs, Angpt2 and Flt1 upregulation was induced by enhanced PKC signaling, without the involvement of canonical Wnt signaling, non-canonical Wnt/Ca2+-mediated activation of NFAT, and non-canonical Wnt/PCP-mediated activation of JNK.