Willis Dissertation Revision Sec1
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

RESTORATION ACTION PLAN MARINA DUNES PRESERVE Marina, California
RESTORATION ACTION PLAN MARINA DUNES PRESERVE Marina, California Prepared for: Monterey Peninsula Regional Park District 4860 Carmel Valley Road Carmel, CA 93923 Prepared by: Burleson Consulting Inc. 1900 Garden Road, Suite 210 Monterey, CA 93940 March 2021 This page intentionally left blank Restoration Action Plan, Marina Dunes Preserve CONTENTS CONTENTS ..........................................................................................................................................i APPENDICES ...................................................................................................................................... ii ACRONYMS AND ABBREVIATIONS ..................................................................................................... iii 1. INTRODUCTION ...................................................................................................................... 1 1.1 Setting ........................................................................................................................................... 1 1.2 Purpose ......................................................................................................................................... 1 1.3 Approach ....................................................................................................................................... 2 2. UPDATED BEST MANAGEMENT PRACTICES .............................................................................. 3 2.1 Weed Eradication and Control ..................................................................................................... -
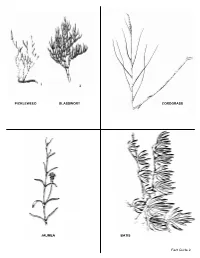
Plant Field Guide
2 PICKLEWEED GLASSWORT CORDGRASS JAUMEA BATIS Field Guide 9 PICKLEWEED Amaranth Family 3 kinds, 2 examples CORDGRASS Grass Family 1 Pickleweed Sarcocornia pacifica Spartina foliosa Glasswort Arthrocnemum subterminalis 2 HABITAT: Growns in the low marsh where the HABITAT: Found throughout the salt marsh. roots are continually bathed in ocean water. APPEARANCE: Stems look like a chain of small APPEARANCE: Look for a tall grass which is pickles. higher than the other plants in the salt marsh. REPRODUCTION: The flowers of all pickleweeds REPRODUCTION: All grasses are wind pollinated. are pollinated by the wind. The small flowers are Look for straw colored spikes of densely packed hard to see because they have no colorful petals flowers. Male flowers will have pollen and the female flowers will show graceful waving stigmas to ADAPTATION TO SALT: Pickleweeds are some of catch the pollen. the many marsh plants that use salt storage (they are accumulators). Also called succulents, these ADAPTATION TO SALT: All the salt marsh plants are swollen with the stored salty water. grasses are salt excreters using special pores to When the salt concentration becomes too high the push out droplets of salty water. Look on the grass cells will die. blades for salt crystals. See sea lavender. ECOLOGICAL RELATIONSHIPS: Frequently the ECOLOGICAL RELATIONSHIPS: Home for the most common plants in the marsh, they provide endangered bird, the Light-footed Clapper Rail. shelter and food for invertebrates. Belding’s A spider lives its whole life inside the blades. Savannah Sparrows build their nests in the Important food for grazing animals. glasswort. BATIS or SALTWORT Saltwort Family Batis maritima HABITAT: Most frequently found in the low marsh. -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

Brassicaceae) in Australia
Cunninghamia Date of Publication: 26/08/2013 A journal of plant ecology for eastern Australia ISSN 0727- 9620 (print) • ISSN 2200 - 405X (Online) Reassessment of the invasion history of two species of Cakile (Brassicaceae) in Australia Roger D. Cousens, Peter K. Ades, Mohsen B. Mesgaran and Sara Ohadi Melbourne School of Land & Environment, The University of Melbourne, Victoria, 3010, AUSTRALIA Abstract: In this paper we revisit the invasion history of two species of Cakile in Australia. Cakile edentula subsp. edentula arrived in the mid 19th Century and spread into coastal strandline habitat from the southeast towards the west and to the north; Cakile maritima arrived in the late 19th Century and has replaced Cakile edentula over much of the range. While Cakile edentula is morphologically quite uniform, the great variation within Cakile maritima has confused field ecologists. Using herbarium records we update previous accounts of the spread of the species and report on field surveys that determined their current geographic overlap in Tasmania and in northern New South Wales/southern Queensland. We examine regional morphological variation within Cakile maritima using the national herbaria collections and variation within new population samples. We support previous interpretations that Cakile maritima has been introduced on more than one occasion from morphologically distinct races, resulting in regional variation within Australia and high variability within populations in the south-east. Western Australian populations appear distinct and probably did not initiate those in the east; we consider that eastern populations are likely to be a mix of Cakile maritima subsp. maritima from the Mediterranean and Cakile maritima subsp. -

State of New York City's Plants 2018
STATE OF NEW YORK CITY’S PLANTS 2018 Daniel Atha & Brian Boom © 2018 The New York Botanical Garden All rights reserved ISBN 978-0-89327-955-4 Center for Conservation Strategy The New York Botanical Garden 2900 Southern Boulevard Bronx, NY 10458 All photos NYBG staff Citation: Atha, D. and B. Boom. 2018. State of New York City’s Plants 2018. Center for Conservation Strategy. The New York Botanical Garden, Bronx, NY. 132 pp. STATE OF NEW YORK CITY’S PLANTS 2018 4 EXECUTIVE SUMMARY 6 INTRODUCTION 10 DOCUMENTING THE CITY’S PLANTS 10 The Flora of New York City 11 Rare Species 14 Focus on Specific Area 16 Botanical Spectacle: Summer Snow 18 CITIZEN SCIENCE 20 THREATS TO THE CITY’S PLANTS 24 NEW YORK STATE PROHIBITED AND REGULATED INVASIVE SPECIES FOUND IN NEW YORK CITY 26 LOOKING AHEAD 27 CONTRIBUTORS AND ACKNOWLEGMENTS 30 LITERATURE CITED 31 APPENDIX Checklist of the Spontaneous Vascular Plants of New York City 32 Ferns and Fern Allies 35 Gymnosperms 36 Nymphaeales and Magnoliids 37 Monocots 67 Dicots 3 EXECUTIVE SUMMARY This report, State of New York City’s Plants 2018, is the first rankings of rare, threatened, endangered, and extinct species of what is envisioned by the Center for Conservation Strategy known from New York City, and based on this compilation of The New York Botanical Garden as annual updates thirteen percent of the City’s flora is imperiled or extinct in New summarizing the status of the spontaneous plant species of the York City. five boroughs of New York City. This year’s report deals with the City’s vascular plants (ferns and fern allies, gymnosperms, We have begun the process of assessing conservation status and flowering plants), but in the future it is planned to phase in at the local level for all species. -

Fort Ord Natural Reserve Plant List
UCSC Fort Ord Natural Reserve Plants Below is the most recently updated plant list for UCSC Fort Ord Natural Reserve. * non-native taxon ? presence in question Listed Species Information: CNPS Listed - as designated by the California Rare Plant Ranks (formerly known as CNPS Lists). More information at http://www.cnps.org/cnps/rareplants/ranking.php Cal IPC Listed - an inventory that categorizes exotic and invasive plants as High, Moderate, or Limited, reflecting the level of each species' negative ecological impact in California. More information at http://www.cal-ipc.org More information about Federal and State threatened and endangered species listings can be found at https://www.fws.gov/endangered/ (US) and http://www.dfg.ca.gov/wildlife/nongame/ t_e_spp/ (CA). FAMILY NAME SCIENTIFIC NAME COMMON NAME LISTED Ferns AZOLLACEAE - Mosquito Fern American water fern, mosquito fern, Family Azolla filiculoides ? Mosquito fern, Pacific mosquitofern DENNSTAEDTIACEAE - Bracken Hairy brackenfern, Western bracken Family Pteridium aquilinum var. pubescens fern DRYOPTERIDACEAE - Shield or California wood fern, Coastal wood wood fern family Dryopteris arguta fern, Shield fern Common horsetail rush, Common horsetail, field horsetail, Field EQUISETACEAE - Horsetail Family Equisetum arvense horsetail Equisetum telmateia ssp. braunii Giant horse tail, Giant horsetail Pentagramma triangularis ssp. PTERIDACEAE - Brake Family triangularis Gold back fern Gymnosperms CUPRESSACEAE - Cypress Family Hesperocyparis macrocarpa Monterey cypress CNPS - 1B.2, Cal IPC -

Mesgaran, M. B., Lewis, M. A., Ades, P. K., Donohue, K
Hybridization can facilitate species invasions, even without enhancing local adaptation Mohsen B. Mesgarana, Mark A. Lewisb, Peter K. Adesc, Kathleen Donohued, Sara Ohadia, Chengjun Lia, and Roger D. Cousensa,1 aSchool of BioSciences, The University of Melbourne, Victoria 3010, Australia; bMathematical and Statistical Sciences, University of Alberta, Edmonton, AB, Canada T6G 2G1; cSchool of Ecosystem and Forest Sciences, The University of Melbourne, Victoria 3010, Australia; and dDepartment of Biology, Duke University, Durham, NC 27708 Edited by Alan Hastings, University of California, Davis, CA, and approved July 5, 2016 (received for review April 6, 2016) The founding population in most new species introductions, or at the low pollinator visitation, or both (8, 9); the term “pollen limitation” leading edge of an ongoing invasion, is likely to be small. Severe isoftenusedasagenerictermwhen the exact mechanism is un- Allee effects—reductions in individual fitness at low population den- clear. In the extreme case of a single arriving adult, population sity—may then result in a failure of the species to colonize, even if persistence would normally be impossible unless the species is ca- the habitat could support a much larger population. Using a simula- pable of asexual reproduction or self-fertilization [“Baker’sLaw” tion model for plant populations that incorporates demography, mat- (10)]. Here, we propose a positive role for hybridization in species ing systems, quantitative genetics, and pollinators, we show that invasions and range expansion, a purely demographic mechanism Allee effects can potentially be overcome by transient hybridization without the requirement for any new adaptation to result. with a resident species or an earlier colonizer. -

Characterization of Mycotoxigenic Alternaria Species Isolated from the Tunisian Halophyte Citation: A
Phytopathologia Mediterranea Firenze University Press The international journal of the www.fupress.com/pm Mediterranean Phytopathological Union Research Paper Characterization of mycotoxigenic Alternaria species isolated from the Tunisian halophyte Citation: A. Chalbi, B. Sghaier-Ham- mami, G. Meca, J.M. Quiles, C. Abdel- Cakile maritima ly, C. Marangi, A.F. Logrieco, A. Moret- ti, M. Masiello (2020) Characterization of mycotoxigenic Alternaria species isolated from the Tunisian halophyte Arbia CHALBI1,2,3, Besma SGHAIER-HAMMAMI1,*, Giuseppe MECA4, Cakile maritima. Phytopathologia Medi- Juan Manuel QUILES4, Chedly ABDELLY1, Carmela MARANGI5, Anto- terranea 59(1): 107-118. doi: 10.14601/ nio F. LOGRIECO3, Antonio MORETTI3, Mario MASIELLO3 Phyto-10720 1 Laboratoire des Plantes Extrêmophiles, Centre de Biotechnologie de Borj-Cédria, BP Accepted: February 7, 2020 901, Hammam-Lif 2050, Tunisia 2 Published: April 30, 2020 Faculty of Sciences of Tunis, University Campus 2092, University of Tunis El Manar, Tunis, Tunisia Copyright: © 2020 A. Chalbi, B. 3 Institute of Sciences of Food Production, Research National Council (ISPA-CNR), Via G. Sghaier-Hammami, G. Meca, J.M. Amendola 122/O, 70126, Bari, Italy Quiles, C. Abdelly, C. Marangi, A.F. 4 Laboratory of Food Toxicology, Department of Preventive Medicine, Nutrition and Food Logrieco, A. Moretti, M. Masiello. This Science Area, Faculty of Pharmacy, University of Valencia Avenida Vicent Andres Estelles is an open access, peer-reviewed s/n, 46100 Burjassot, Valencia, Spain article published by Firenze Univer- 5 sity Press (http://www.fupress.com/pm) Institute for Applied Mathematics “M. Picone”, Via G. Amendola 122/D, 70126, Bari, and distributed under the terms of the Italy Creative Commons Attribution License, * Corresponding author: [email protected] which permits unrestricted use, distri- bution, and reproduction in any medi- um, provided the original author and Summary. -

The Brassicaceae of Ohio
THE BRASSICACEJE OF OHIO. EMMA E. LAUGHLIN. Brassicaceae. Mustard Family. Herbs, with watery sap of a pungent taste, not poisonous; with alternate, exstipulate leaves, usually large at the base of the stem and intergrading in form to the top of the stem. Flowers hypogynous, bisporangiate, usually isobilateral, appear- ing actinomorphic, regular, usually with glands, in racemes, short at first and elongating, or in corymbs; calyx of 4 sepals, decidu- ous, rarely persistent; corolla choripetalous, tetramerous, cruci- form; stamens 6, tetradynamous, rarely 4 or 2; ovulary com- pound, bilocular, the parietal placentae connected by a thin septum from which the valves separate when ripe; ovules 2 to several, campylotropous; fruit a silique if longer than broad, or a silicle if short, generally with 2 cavities, sometimes uni- locular, dehiscent or in a few genera indehiscent; endosperm scanty; cotyledons accumbent, incumbent or conduplicate. SYNOPSIS. I. Pod usually not more than twice as long as wide (a silicle); cotyledons accum- bent or incumbent. A. Pods more or less flattened parallel to the broad partition, dehiscent; cotyledons accumbent; leaves not lobed. 1. Pubescence stellate or of forked hairs. Berteroa, Koniga, Alyssum, Draba. 2. Pubescence of simple hairs or wanting; pods very broad and flat; leaves opposite. LUNARIE^E. Lunaria. B. Pods flattened at right angles to the partition or not flattened. 1. Pubescence of forked hairs; cotyledons incumbent. CAMELINE^E. Camelina, Bursa, Neslia. 2. Pubescence of simple hairs or wanting. a. Pod scarcely or not at all flattened; cotyledous accumbent. COCHLEARIE^E. Armoracia, Neobeckia, Sisymbrium, Radicula. b. Pods strongly flattened at right angles to the narrow partition. -

Vascular Plants of the Coastal Dunes of Humboldt County, California
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 4-2019 Vascular Plants of the Coastal Dunes of Humboldt County, California James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: https://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Vascular Plants of the Coastal Dunes of Humboldt County, California" (2019). Botanical Studies. 41. https://digitalcommons.humboldt.edu/botany_jps/41 This Flora of Northwest California-Checklists of Local Sites is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. VASCULAR PLANTS OF THE COASTAL DUNES OF HUMBOLDT COUNTY, CALIFORNIA Compiled by James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State University Arcata, California Ninth Edition • August 2019 Amaryllidaceae — Onion or Amaryllis Family F E R N S Allium unifolium •One-leaved onion Dennstaedtiaceae — Bracken Fern Family Anacardiaceae — Cashew Family Pteridium aquilinum var. pubescens • Bracken fern Toxicodendron diversilobum • Poison-oak Ophioglossaceae — Adder’s-tongue Family Apocynaceae — Dogbane or Milkweed Family Botrychium multifidum • Leathery -

Vascular Plants of Humboldt Bay's Dunes and Wetlands Published by U.S
Vascular Plants of Humboldt Bay's Dunes and Wetlands Published by U.S. Fish and Wildlife Service G. Leppig and A. Pickart and California Department of Fish Game Release 4.0 June 2014* www.fws.gov/refuge/humboldt_bay/ Habitat- Habitat - Occurs on Species Status Occurs within Synonyms Common name specific broad Lanphere- Jepson Manual (2012) (see codes at end) refuge (see codes at end) (see codes at end) Ma-le'l Units UD PW EW Adoxaceae Sambucus racemosa L. red elderberry RF, CDF, FS X X N X X Aizoaceae Carpobrotus chilensis (Molina) sea fig DM X E X X N.E. Br. Carpobrotus edulis ( L.) N.E. Br. Iceplant DM X E, I X Alismataceae lanceleaf water Alisma lanceolatum With. FM X E plantain northern water Alisma triviale Pursh FM X N plantain Alliaceae three-cornered Allium triquetrum L. FS, FM, DM X X E leek Allium unifolium Kellogg one-leaf onion CDF X N X X Amaryllidaceae Amaryllis belladonna L. belladonna lily DS, AW X X E Narcissus pseudonarcissus L. daffodil AW, DS, SW X X E X Anacardiaceae Toxicodendron diversilobum Torrey poison oak CDF, RF X X N X X & A. Gray (E. Greene) Apiaceae Angelica lucida L. seacoast angelica BM X X N, C X X Anthriscus caucalis M. Bieb bur chevril DM X E Cicuta douglasii (DC.) J. Coulter & western water FM X N Rose hemlock Conium maculatum L. poison hemlock RF, AW X I X Daucus carota L. Queen Anne's lace AW, DM X X I X American wild Daucus pusillus Michaux DM, SW X X N X X carrot Foeniculum vulgare Miller sweet fennel AW, FM, SW X X I X Glehnia littoralis (A. -

The Germination Niche of Coastal Dune Species As Related to Their Occurrence Along a Sea–Inland Gradient
Received: 16 September 2019 | Revised: 18 March 2020 | Accepted: 6 April 2020 DOI: 10.1111/jvs.12899 SPECIAL FEATURE: DISPERSAL AND ESTABLISHMENT Journal of Vegetation Science The germination niche of coastal dune species as related to their occurrence along a sea–inland gradient Silvia Del Vecchio1 | Edy Fantinato1 | Mauro Roscini1 | Alicia T. R. Acosta2 | Gianluigi Bacchetta3 | Gabriella Buffa1 1Department of Environmental Sciences, Informatics and Statistics, Ca’ Foscari Abstract University of Venice, Venice, Italy Aims: The early phases in the life cycle of a plant are the bottleneck for success- 2 Department of Sciences, University of ful species establishment thereby affecting population dynamics and distribution. In Rome 3, Rome, Italy 3Sardinian Germplasm Bank (BG-SAR), coastal environments, the spatial pattern of plant communities (i.e. vegetation zona- Hortus Botanicus Karalitanus (HBK), tion) follows the ecological gradient of abiotic stress changing with the distance from University of Cagliari, Cagliari, Italy the sea. This pattern has been mainly explained based on the adaptation and toler- Correspondence ance to the abiotic stress of adult plants. However, the adult niche may considerably Silvia Del Vecchio, Department of Environmental Sciences, Informatics and differ from the germination niche of a plant species. The aim of this work was to Statistics, Ca’ Foscari University of Venice, investigate to what extent abiotic factors (specifically salinity, temperature, nitrogen Via Torino 155, 30172 Venice, Italy Email: [email protected] and their interactions) constrain seed germination along the sea–inland gradient. Location: Latium coast (Central Italy). Co-ordinating Editor: Judit Sonkoly Methods: Germination tests were performed on seeds of focal species of three dif- This article is a part of the Special Feature ferent plant communities which establish at increasing distances from the coast- Plant dispersal and establishment as drivers line: Cakile maritima subsp.