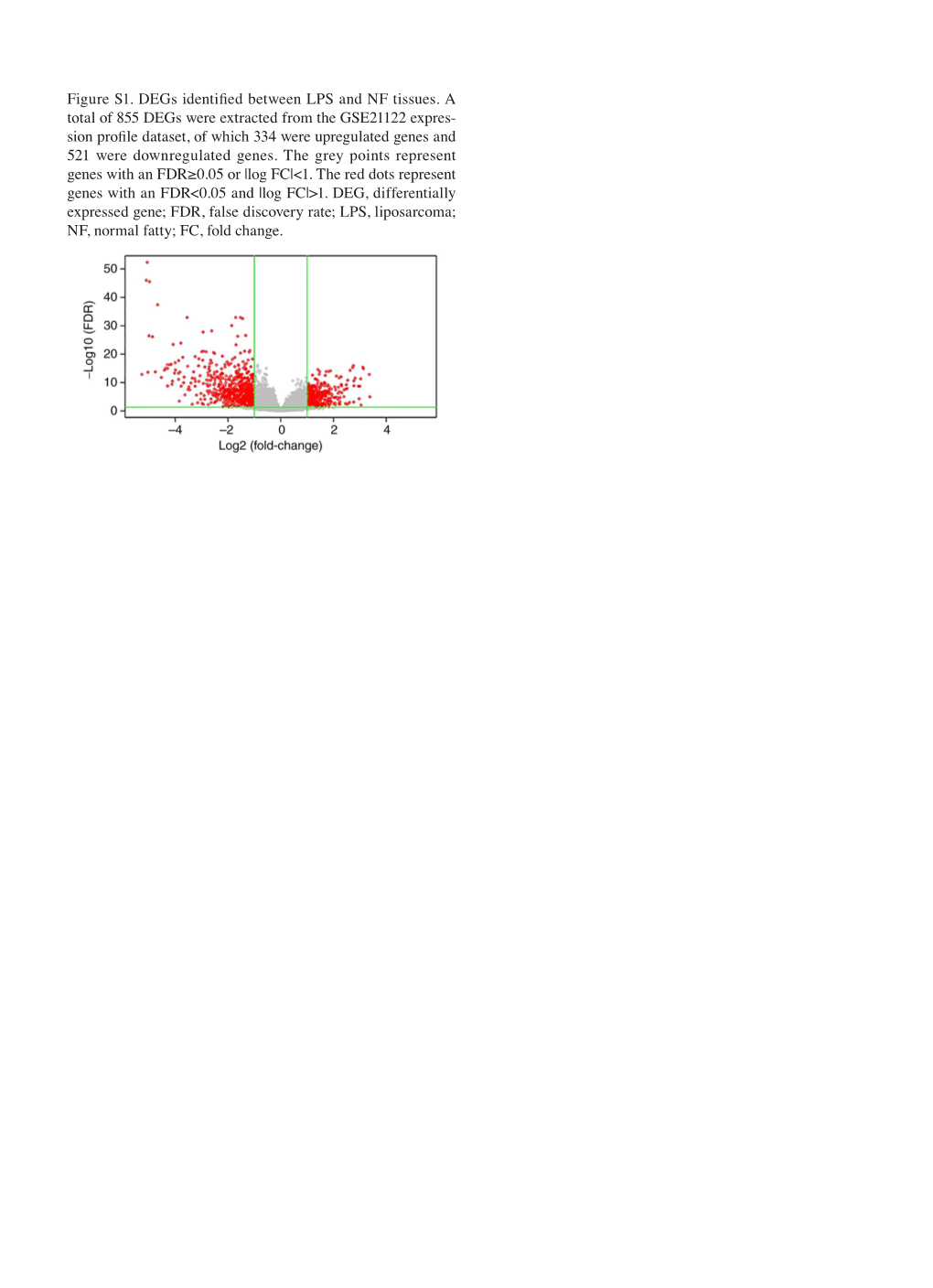
Figure S1. Degs Identified Between LPS and NF Tissues
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Kinesin-14 Motor Protein KIFC1 Participates in DNA Synthesis and Chromatin Maintenance Ya-Lan Wei1 and Wan-Xi Yang 1
Wei and Yang Cell Death and Disease (2019) 10:402 https://doi.org/10.1038/s41419-019-1619-9 Cell Death & Disease ARTICLE Open Access Kinesin-14 motor protein KIFC1 participates in DNA synthesis and chromatin maintenance Ya-Lan Wei1 and Wan-Xi Yang 1 Abstract The nuclear localization signal (NLS) in kinesin-14 KIFC1 is associated with nuclear importins and Ran gradient, but detailed mechanism remains unknown. In this study, we found that KIFC1 proteins have specific transport characteristics during cell cycle. In the absence of KIFC1, cell cycle kinetics decrease significantly with a prolonged S phase. After KIFC1 overexpression, the duration of S phase becomes shorten. KIFC1 may transport the recombinant/ replicate-related proteins into the nucleus, meanwhile avoiding excessive KIFC1 in the cytoplasm, which results in aberrant microtubule bundling. Interestingly, the deletion of kifc1 in human cells results in a higher ratio of aberrant nuclear membrane, and the degradation of lamin B and lamin A/C. We also found that kifc1 deletion leads to defects in metaphase mitotic spindle assembly, and then results in chromosome structural abnormality. The kifc1-/- cells finally form micronuclei in daughter cells, and results in aneuploidy and chromosome loss in cell cycle. In this study, we demonstrate that kinesin-14 KIFC1 proteins involve in regulating DNA synthesis in S phase, and chromatin maintenance in mitosis, and maintain cell growth in a nuclear transport-independent way. 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; Introduction KIFC1 mainly cluster the spindles involving in chromo- Kinesin-14 KIFC1 transports various cargos along the some alignment and segregation. While chromokinesins microtubule to the minus ends1. -

Ran Activation Assay Kit
Product Manual Ran Activation Assay Kit Catalog Number STA-409 20 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Small GTP-binding proteins (or GTPases) are a family of proteins that serve as molecular regulators in signaling transduction pathways. Ran, a 25 kDa protein of the Ras superfamily, regulates a variety of biological response pathways that include DNA synthesis, cell cycle progression, and translocation of RNA/proteins through the nuclear pore complex. Like other small GTPases, Ran regulates molecular events by cycling between an inactive GDP-bound form and an active GTP-bound form. In its active (GTP-bound) state, Ran binds specifically to RanBP1 to control downstream signaling cascades. Cell Biolabs’ Ran Activation Assay Kit utilizes RanBP1 Agarose beads to selectively isolate and pull- down the active form of Ran from purified samples or endogenous lysates. Subsequently, the precipitated GTP-Ran is detected by western blot analysis using an anti-Ran antibody. Cell Biolabs’ Ran Activation Assay Kit provides a simple and fast tool to monitor the activation of Ran. The kit includes easily identifiable RanBP1 Agarose beads (see Figure 1), pink in color, and a GTPase Immunoblot Positive Control for quick Ran identification. Each kit provides sufficient quantities to perform 20 assays. Figure 1: RanBP1 Agarose beads, in color, are easy to visualize, minimizing potential loss during washes and aspirations. 2 Assay Principle Related Products 1. STA-400: Pan-Ras Activation Assay Kit 2. STA-400-H: H-Ras Activation Assay Kit 3. STA-400-K: K-Ras Activation Assay Kit 4. STA-400-N: N-Ras Activation Assay Kit 5. -

Roles and Mechanisms of Kinesin-6 KIF20A in Spindle Organization During Cell Division T ⁎ Wen-Da Wu, Kai-Wei Yu, Ning Zhong, Yu Xiao, Zhen-Yu She
European Journal of Cell Biology 98 (2019) 74–80 Contents lists available at ScienceDirect European Journal of Cell Biology journal homepage: www.elsevier.com/locate/ejcb Review Roles and mechanisms of Kinesin-6 KIF20A in spindle organization during cell division T ⁎ Wen-Da Wu, Kai-Wei Yu, Ning Zhong, Yu Xiao, Zhen-Yu She Department of Cell Biology and Genetics/Center for Cell and Developmental Biology, The School of Basic Medical Sciences, Fujian Medical University, Fuzhou, Fujian 350108, China ARTICLE INFO ABSTRACT Keywords: Mitotic kinesin is crucial for spindle assembly and chromosome segregation in cell division. KIF20A/MKlp2, a Kinesin-6 member of kinesin-6 subfamily, plays important roles in the central spindle organization at anaphase and cy- KIF20A tokinesis. In this review, we briefly introduce the discovery and classification of kinesin-6 motors in model Microtubule organisms, and summarize the biochemical features and mechanics of KIF20A proteins. We emphasize the Anaphase complicated interactions of KIF20A with partner proteins, including MKlp1, Plk1 and Rab6. Particularly, we Spindle assembly highlight the regulation of Cdk1 and chromosomal passenger complex on kinesin-6 KIF20A at late stage of Mitosis mitosis. We summarized the multiple functions of KIF20A in central spindle assembly and the formation of cleavage furrow in both mitosis and meiosis. In addition, we conclude the expression patterns of KIF20A in tumorigenesis and its applications in tumor therapy. 1. Introduction kinesin superfamily proteins (Miki et al., 2005). Kinesin-6 subfamily is comprised of KIF20A (Lawrence et al., 2004), KIF20B (MPP1) Kinesin superfamily proteins (KIFs) are molecular motors that (Kamimoto et al., 2001; Matsumoto-Taniura et al., 1996; Westendorf mediate the transport of various cargos, including the newly synthe- et al., 1994) and MKlp1 (Lawrence et al., 2004; Nislow et al., 1990; sized protein complexes, vesicles and mRNAs along the microtubule Sellitto and Kuriyama, 1988). -

Rhob Controls Endothelial Barrier Recovery by Inhibiting Rac1 Trafficking to the Cell Border
JCB: Article RhoB controls endothelial barrier recovery by inhibiting Rac1 trafficking to the cell border Beatriz Marcos‑Ramiro,1 Diego García‑Weber,1 Susana Barroso,1 Jorge Feito,2 María C. Ortega,1 Eva Cernuda‑Morollón,3 Natalia Reglero‑Real,1 Laura Fernández‑Martín,1 Maria C. Durán,4 Miguel A. Alonso,1 Isabel Correas,1 Susan Cox,5 Anne J. Ridley,5 and Jaime Millán1 1Centro de Biología Molecular Severo Ochoa, Consejo Superior de Investigaciones Cientificas, Universidad Autónoma de Madrid, 28049 Madrid, Spain 2Servicio de Anatomía Patológica, Hospital Universitario de Salamanca, 37007 Salamanca, Spain 3Neurology Department, Hospital Universitario Central de Asturias, 33011 Oviedo, Spain 4Biomedicine, Biotechnology and Public Health Department, University of Cadiz, 11519 Cadiz, Spain 5Randall Division of Cell and Molecular Biophysics, King’s College London, SE1 1UL London, England, UK Endothelial barrier dysfunction underlies chronic inflammatory diseases. In searching for new proteins essential to the human endothelial inflammatory response, we have found that the endosomal GTPase RhoB is up-regulated in response to inflammatory cytokines and expressed in the endothelium of some chronically inflamed tissues. We show that al- though RhoB and the related RhoA and RhoC play additive and redundant roles in various aspects of endothelial barrier function, RhoB specifically inhibits barrier restoration after acute cell contraction by preventing plasma membrane ex- tension. During barrier restoration, RhoB trafficking is induced between vesicles containing RhoB nanoclusters and plasma membrane protrusions. The Rho GTPase Rac1 controls membrane spreading and stabilizes endothelial barriers. We show that RhoB colocalizes with Rac1 in endosomes and inhibits Rac1 activity and trafficking to the cell border during barrier recovery. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Prox1regulates the Subtype-Specific Development of Caudal Ganglionic
The Journal of Neuroscience, September 16, 2015 • 35(37):12869–12889 • 12869 Development/Plasticity/Repair Prox1 Regulates the Subtype-Specific Development of Caudal Ganglionic Eminence-Derived GABAergic Cortical Interneurons X Goichi Miyoshi,1 Allison Young,1 Timothy Petros,1 Theofanis Karayannis,1 Melissa McKenzie Chang,1 Alfonso Lavado,2 Tomohiko Iwano,3 Miho Nakajima,4 Hiroki Taniguchi,5 Z. Josh Huang,5 XNathaniel Heintz,4 Guillermo Oliver,2 Fumio Matsuzaki,3 Robert P. Machold,1 and Gord Fishell1 1Department of Neuroscience and Physiology, NYU Neuroscience Institute, Smilow Research Center, New York University School of Medicine, New York, New York 10016, 2Department of Genetics & Tumor Cell Biology, St. Jude Children’s Research Hospital, Memphis, Tennessee 38105, 3Laboratory for Cell Asymmetry, RIKEN Center for Developmental Biology, Kobe 650-0047, Japan, 4Laboratory of Molecular Biology, Howard Hughes Medical Institute, GENSAT Project, The Rockefeller University, New York, New York 10065, and 5Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724 Neurogliaform (RELNϩ) and bipolar (VIPϩ) GABAergic interneurons of the mammalian cerebral cortex provide critical inhibition locally within the superficial layers. While these subtypes are known to originate from the embryonic caudal ganglionic eminence (CGE), the specific genetic programs that direct their positioning, maturation, and integration into the cortical network have not been eluci- dated. Here, we report that in mice expression of the transcription factor Prox1 is selectively maintained in postmitotic CGE-derived cortical interneuron precursors and that loss of Prox1 impairs the integration of these cells into superficial layers. Moreover, Prox1 differentially regulates the postnatal maturation of each specific subtype originating from the CGE (RELN, Calb2/VIP, and VIP). -

Molecular Profiling of Peripheral Blood Is Associated with Circulating Tumor Cells Content and Poor Survival in Metastatic Castration-Resistant Prostate Cancer
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No. 12 Molecular profiling of peripheral blood is associated with circulating tumor cells content and poor survival in metastatic castration-resistant prostate cancer Mercedes Marín-Aguilera1, Òscar Reig1,2, Juan José Lozano3, Natalia Jiménez1, Susana García-Recio1,4, Nadina Erill5, Lydia Gaba2, Andrea Tagliapietra2, Vanesa Ortega2, Gemma Carrera6, Anna Colomer5, Pedro Gascón4 and Begoña Mellado1,2 1 Translational Genomics Group and Targeted Therapeutics in Solid Tumors Group, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Spain 2 Medical Oncology Department, Hospital Clínic, Barcelona, Spain 3 Bioinformatics Platform Department, Centro de Investigación Biomédica en Red en el Área temática de Enfermedades Hepáticas y Digestivas (CIBEREHD), Hospital Clínic, Barcelona, Spain 4 Laboratory of Translational Oncology, Fundació Clínic per a la Recerca Biomèdica, Barcelona, Spain 5 Althia, Barcelona, Spain 6 Medical Oncology Department, Hospital Plató, Barcelona, Spain Correspondence to: Begoña Mellado, email: [email protected] Keywords: circulating tumor cells, peripheral blood, microarrays, cell search system Received: January 22, 2015 Accepted: February 14, 2015 Published: March 12, 2015 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT The enumeration of circulating -

A Flexible Microfluidic System for Single-Cell Transcriptome Profiling
www.nature.com/scientificreports OPEN A fexible microfuidic system for single‑cell transcriptome profling elucidates phased transcriptional regulators of cell cycle Karen Davey1,7, Daniel Wong2,7, Filip Konopacki2, Eugene Kwa1, Tony Ly3, Heike Fiegler2 & Christopher R. Sibley 1,4,5,6* Single cell transcriptome profling has emerged as a breakthrough technology for the high‑resolution understanding of complex cellular systems. Here we report a fexible, cost‑efective and user‑ friendly droplet‑based microfuidics system, called the Nadia Instrument, that can allow 3′ mRNA capture of ~ 50,000 single cells or individual nuclei in a single run. The precise pressure‑based system demonstrates highly reproducible droplet size, low doublet rates and high mRNA capture efciencies that compare favorably in the feld. Moreover, when combined with the Nadia Innovate, the system can be transformed into an adaptable setup that enables use of diferent bufers and barcoded bead confgurations to facilitate diverse applications. Finally, by 3′ mRNA profling asynchronous human and mouse cells at diferent phases of the cell cycle, we demonstrate the system’s ability to readily distinguish distinct cell populations and infer underlying transcriptional regulatory networks. Notably this provided supportive evidence for multiple transcription factors that had little or no known link to the cell cycle (e.g. DRAP1, ZKSCAN1 and CEBPZ). In summary, the Nadia platform represents a promising and fexible technology for future transcriptomic studies, and other related applications, at cell resolution. Single cell transcriptome profling has recently emerged as a breakthrough technology for understanding how cellular heterogeneity contributes to complex biological systems. Indeed, cultured cells, microorganisms, biopsies, blood and other tissues can be rapidly profled for quantifcation of gene expression at cell resolution. -

Wnt/Β-Catenin and Sex Hormone Signaling in Endometrial Homeostasis and Cancer
www.impactjournals.com/oncotarget/ Oncotarget, November, Vol.1, No 7 Wnt/Β-Catenin and Sex Hormone Signaling in Endometrial Homeostasis and Cancer Yongyi Wang1,2, Marten van der Zee1,2, Riccardo Fodde2, Leen J Blok1 1 Department of Obstetrics & Gynaecology, Josephine Nefkens Institute, Erasmus University Medical Center Rotterdam, PO Box 2040, 3000 CA Rotterdam, The Netherlands 2 Departments of Pathology, Josephine Nefkens Institute, Erasmus University Medical Center Rotterdam, PO Box 2040, 3000 CA Rotterdam, The Netherlands Correspondence to: Leen J Blok, e-mail: [email protected] Keywords: Wnt/β-catenin, estradiol, progesterone, endometrium Received: September 30, 2010, Accepted: October 11, 2010, Published: October 12, 2010 Copyright: © Wang et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT: A delicate balance between estrogen and progestagen signaling underlies proper functioning of the female reproductive tract and, in particular, the monthly re- and degenerative phases characteristic of the menstrual cycle. Here, we propose that the canonical Wnt/β-catenin signaling pathway may underlie this finely tuned hormonal equilibrium in endometrial homeostasis and, upon its constitutive activation, lead to neoplastic transformation of the endometrium. During the menstrual cycle, estradiol will enhance Wnt/β-catenin signaling in the proliferative phase, while progesterone inhibits Wnt/β-catenin signaling, thus restraining estrogens’ proliferative actions, during the secretory phase. In case of enhanced or unopposed estrogen signaling, constitutive activation of Wnt/β-catenin signaling will trigger endometrial hyperplasia, which may develop further into endometrial cancer. -

Screening and Identification of Hub Genes in Bladder Cancer by Bioinformatics Analysis and KIF11 Is a Potential Prognostic Biomarker
ONCOLOGY LETTERS 21: 205, 2021 Screening and identification of hub genes in bladder cancer by bioinformatics analysis and KIF11 is a potential prognostic biomarker XIAO‑CONG MO1,2*, ZI‑TONG ZHANG1,3*, MENG‑JIA SONG1,2, ZI‑QI ZHOU1,2, JIAN‑XIONG ZENG1,2, YU‑FEI DU1,2, FENG‑ZE SUN1,2, JIE‑YING YANG1,2, JUN‑YI HE1,2, YUE HUANG1,2, JIAN‑CHUAN XIA1,2 and DE‑SHENG WENG1,2 1State Key Laboratory of Oncology in South China, Collaborative Innovation Centre for Cancer Medicine; 2Department of Biotherapy, Sun Yat‑Sen University Cancer Center; 3Department of Radiation Oncology, Sun Yat‑Sen University Cancer Center, Guangzhou, Guangdong 510060, P.R. China Received July 31, 2020; Accepted December 18, 2020 DOI: 10.3892/ol.2021.12466 Abstract. Bladder cancer (BC) is the ninth most common immunohistochemistry and western blotting. In summary, lethal malignancy worldwide. Great efforts have been devoted KIF11 was significantly upregulated in BC and might act as to clarify the pathogenesis of BC, but the underlying molecular a potential prognostic biomarker. The present identification mechanisms remain unclear. To screen for the genes associated of DEGs and hub genes in BC may provide novel insight for with the progression and carcinogenesis of BC, three datasets investigating the molecular mechanisms of BC. were obtained from the Gene Expression Omnibus. A total of 37 tumor and 16 non‑cancerous samples were analyzed to Introduction identify differentially expressed genes (DEGs). Subsequently, 141 genes were identified, including 55 upregulated and Bladder cancer (BC) is the ninth most common malignancy 86 downregulated genes. The protein‑protein interaction worldwide with substantial morbidity and mortality. -

Expression Signatures of the Lipid-Based Akt Inhibitors Phosphatidylinositol Ether Lipid Analogues in NSCLC Cells
Published OnlineFirst May 6, 2011; DOI: 10.1158/1535-7163.MCT-10-1028 Molecular Cancer Therapeutic Discovery Therapeutics Expression Signatures of the Lipid-Based Akt Inhibitors Phosphatidylinositol Ether Lipid Analogues in NSCLC Cells Chunyu Zhang1, Abdel G. Elkahloun2, Hongling Liao3, Shannon Delaney1, Barbara Saber1, Betsy Morrow1, George C. Prendergast4, M. Christine Hollander1, Joell J. Gills1, and Phillip A. Dennis1 Abstract Activation of the serine/threonine kinase Akt contributes to the formation, maintenance, and therapeutic resistance of cancer, which is driving development of compounds that inhibit Akt. Phosphatidylinositol ether lipid analogues (PIA) are analogues of the products of phosphoinositide-3-kinase (PI3K) that inhibit Akt activation, translocation, and the proliferation of a broad spectrum of cancer cell types. To gain insight into the mechanism of PIAs, time-dependent transcriptional profiling of five active PIAs and the PI3K inhibitor LY294002 (LY) was conducted in non–small cell lung carcinoma cells using high-density oligonucleotide arrays. Gene ontology analysis revealed that genes involved in apoptosis, wounding response, and angiogen- esis were upregulated by PIAs, whereas genes involved in DNA replication, repair, and mitosis were suppressed. Genes that exhibited early differential expression were partitioned into three groups; those induced by PIAs only (DUSP1, KLF6, CENTD2, BHLHB2, and PREX1), those commonly induced by PIAs and LY (TRIB1, KLF2, RHOB, and CDKN1A), and those commonly suppressed by PIAs and LY (IGFBP3, PCNA, PRIM1, MCM3, and HSPA1B). Increased expression of the tumor suppressors RHOB (RhoB), KLF6 (COPEB), and CDKN1A (p21Cip1/Waf1) was validated as an Akt-independent effect that contributed to PIA-induced cytotoxicity. Despite some overlap with LY, active PIAs have a distinct expression signature that contributes to their enhanced cytotoxicity. -

Characterization of KIF20A As a Prognostic Biomarker and Therapeutic Target for Different Subtypes of Breast Cancer
INTERNATIONAL JOURNAL OF ONCOLOGY 57: 277-288, 2020 Characterization of KIF20A as a prognostic biomarker and therapeutic target for different subtypes of breast cancer MASAKO NAKAMURA1,2, ATSUSHI TAKANO1-3, PHUNG MANH THANG1-3, BAYARBAT TSEVEGJAV1,2, MING ZHU1,2, TOMOYUKI YOKOSE4, TOSHINARI YAMASHITA5, YOHEI MIYAGI6 and YATARO DAIGO1-3 1Department of Medical Oncology and Cancer Center; 2Center for Advanced Medicine against Cancer, Shiga University of Medical Science, Otsu, Shiga 520-2192; 3Center for Antibody and Vaccine Therapy, Research Hospital, Institute of Medical Science, The University of Tokyo, Tokyo 108-8639; Departments of 4Pathology, and 5Breast and Endocrine Surgery, Kanagawa Cancer Center; 6Molecular Pathology and Genetics Division, Kanagawa Cancer Center Research Institute, Yokohama, Kanagawa 241-8515, Japan Received November 11, 2019; Accepted March 6, 2020 DOI: 10.3892/ijo.2020.5060 Abstract. The aim of the present study was to identify novel Introduction prognostic biomarkers and therapeutic targets for breast cancer; thus, genes that are frequently overexpressed in Breast, lung and bronchial and colorectal cancer are the three several types of breast cancer were screened. Kinesin family most commonly diagnosed types of cancer among women and member 20A (KIF20A) was identified as a candidate molecule represent 50% of all cases. However, breast cancer alone is during this process. Immunohistochemical staining performed expected to account for 30% of all new cancer cases diagnosed using tissue microarrays from 257 samples of different breast among women (1). Breast cancer is a collection of conditions cancer subtypes revealed that KIF20A was expressed in 195 that have different biological properties and can arise from (75.9%) of these samples, whereas it was seldom expressed various genetic abnormalities.