Mouse Genetic Background Influences Whether Hrasg12v Expression Plus Cdkn2a Knockdown Causes Angiosarcoma Or Undifferentiated Pleomorphic Sarcoma
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Glomus Tumor in the Floor of the Mouth: a Case Report and Review of the Literature Haixiao Zou1,2, Li Song1, Mengqi Jia2,3, Li Wang4 and Yanfang Sun2,3*
Zou et al. World Journal of Surgical Oncology (2018) 16:201 https://doi.org/10.1186/s12957-018-1503-6 CASEREPORT Open Access Glomus tumor in the floor of the mouth: a case report and review of the literature Haixiao Zou1,2, Li Song1, Mengqi Jia2,3, Li Wang4 and Yanfang Sun2,3* Abstract Background: Glomus tumors are rare benign neoplasms that usually occur in the upper and lower extremities. Oral cavity involvement is exceptionally rare, with only a few cases reported to date. Case presentation: A 24-year-old woman with complaints of swelling in the left floor of her mouth for 6 months was referred to our institution. Her swallowing function was slightly affected; however, she did not have pain or tongue paralysis. Enhanced computed tomography revealed a 2.8 × 1.8 × 2.1 cm-sized well-defined, solid, heterogeneous nodule above the mylohyoid muscle. The mandible appeared to be uninvolved. The patient underwent surgery via an intraoral approach; histopathological examination revealed a glomus tumor. The patient has had no evidence of recurrence over 4 years of follow-up. Conclusions: Glomus tumors should be considered when patients present with painless nodules in the floor of the mouth. Keywords: Glomus tumor, Floor of mouth, Oral surgery Background Case presentation Theglomusbodyisaspecialarteriovenousanasto- A 24-year-old woman with a 6-month history of swelling mosisandfunctionsinthermalregulation.Glomustu- in the left floor of her mouth was referred to our institu- mors are rare, benign, mesenchymal tumors that tion. Although she experienced slight difficulty in swal- originate from modified smooth muscle cells of the lowing, she did not experience pain or tongue paralysis. -

Angiosarcomas and Other Sarcomas of Endothelial Origin
Angiosarcomas and Other Sarcomas of Endothelial Origin a,b a Angela Cioffi, MD , Sonia Reichert, MD , c a,b,d, Cristina R. Antonescu, MD , Robert G. Maki, MD, PhD, FACP * KEYWORDS Angiosarcoma Epithelioid hemangioendothelioma Vascular sarcoma Kaposi sarcoma VEGF KDR FLT4 Translocation Organ transplant KEY POINTS Vascular sarcomas are rare and collectively affect fewer than 600 people a year in the United States (incidence approximately 2/million). Because angiosarcomas, hemangioendotheliomas, and other vascular tumors have unique embryonal derivation, it is not surprising that they have a unique sensitivity pattern to chemotherapy agents. Surgery, when possible, remains the primary treatment for angiosarcomas. Adjuvant radiation for primary disease seems prudent for at least some angiosarcoma, given the high local-regional recurrence rate of these tumors. Angiosarcomas also have a high rate of metastasis, but it is not clear that adjuvant chemotherapy improves survival. Epithelioid hemangioendothelioma is a unique form of sarcoma often presenting as multi- focal disease. Most patients can do well with observation alone, although a fraction of pa- tients have more aggressive disease and have difficulties in both local control and metastatic disease. Continued Disclosures: R.G. Maki receives clinical research support from Morphotek/Eisai, Ziopharm, and Imclone/Lilly. He has also consulted for Eisai, Morphotek/Eisai, Imclone/Lilly, Taiho, Glaxo- SmithKline, Merck, Champions Biotechnology, and Pfizer. He has received speaker’s fees from Novartis. He is an unpaid consultant for the Sarcoma Foundation of America, SARC: Sarcoma Alliance for Research through Collaboration, n-of-one, and 23 & me. C.R. Antonescu, A. Cioffi, and S. Reichert report no conflicts. -

Malignant Vascular Tumors&Mdash
Modern Pathology (2014) 27, S30–S38 S30 & 2014 USCAP, Inc All rights reserved 0893-3952/14 $32.00 Malignant vascular tumors—an update Cristina Antonescu Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, NY, USA Although benign hemangiomas are among the most common diagnoses amid connective tissue tumors, sarcomas showing endothelial differentiation (ie, angiosarcoma and epithelioid hemangioendothelioma) represent under 1% of all sarcoma diagnoses, and thus it is likely that fewer than 500 people in the United States are affected each year. Differential diagnosis of malignant vascular tumors can be often quite challenging, either at the low end of the spectrum, distinguishing an epithelioid hemangioendothelioma from an epithelioid hemangioma, or at the high-grade end of the spectrum, between an angiosarcoma and a malignant epithelioid hemangioendothelioma. Within this differential diagnosis both clinico-radiological features (ie, size and multifocality) and immunohistochemical markers (ie, expression of endothelial markers) are often similar and cannot distinguish between benign and malignant vascular lesions. Molecular ancillary tests have long been needed for a more objective diagnosis and classification of malignant vascular tumors, particularly within the epithelioid phenotype. As significant advances have been recently made in understanding the genetic signatures of vascular tumors, this review will take the opportunity to provide a detailed update on these findings. Specifically, this article will focus on -

Benign Lymphangioendothelioma Manifested Clinically As Actinic Keratosis James A
Benign Lymphangioendothelioma Manifested Clinically as Actinic Keratosis James A. Yiannias, MD, Scottsdale, Arizona R.K. Winkelmann, MD, PhD, Scottsdale, Arizona Benign lymphangioendothelioma is an acquired lymphangiectatic lesion that must be recognized and differentiated from angiosarcoma, early Kaposi’s sarcoma, and lymphangioma circum- scriptum. We report the case of a 68-year-old woman with the clinical presentation of a possible actinic keratosis and the typical histologic findings of benign lymphangioendothelioma and an overly- ing actinic keratosis. enign lymphangioendothelioma is a recently de- scribed acquired lymphangiectatic lesion. Clin- ically, it usually appears as a dull pink to red- B 1 dish brown macule or plaque. We report a case in which the clinical presentation was possible actinic FIGURE 1. Benign lymphangioendothelioma at extensor keratosis, with typical histologic findings of benign forearm after punch biopsy. lymphangioendothelioma and an overlying pig- mented actinic keratosis. Recognition of this entity is vital because the histologic differential diagnosis cally, a pigmented actinic keratosis or lentigo was sus- includes angiosarcoma, early Kaposi’s sarcoma, and pected. A punch biopsy specimen showed delicate, lymphangioma circumscriptum. thin-walled, endothelium-lined spaces and clefts in the upper dermis, with an overlying pigmented Case Report actinic keratosis (Figure 2). These vascular channels A 68-year-old white woman in generally good health ran parallel to the epidermis and contained no or few presented with a small, light brown patch on the erythrocytes in their lumina. The endothelial cells extensor surface of her right forearm that grew radi- outlined collagen bundles. Furthermore, there was no ally over a 2-year period. The lesion was asymptom- erythrocyte extravasation, hemosiderin deposition, or atic, but the patient was concerned about cosmesis significant inflammation, and no abnormal muscular and requested that it be removed. -

Hepatic Angiosarcoma Masquerading As Hemangioma
Hepatic Angiosarcoma Masquerading as Hemangioma: A CASO CLÍNICO Challenging Differential Diagnosis Angiosarcoma Hepático e Hemangioma: Um Diagnóstico Diferencial Desafiante Ana Rita GARCIA1, João RIBEIRO1, Helena GERVÁSIO1, Francisco Castro e SOUSA2,3 Acta Med Port 2017 Oct;30(10):750-753 ▪ https://doi.org/10.20344/amp.8593 ABSTRACT Hemangiomas are usually diagnosed based on ultrasound findings. The presence of symptoms, rapid growth or atipical imagiological findings should make us consider other diagnoses, including malignant tumors such as angiosarcomas. We describe the case of a previously healthy 46-year-old female without a history of exposure to carcinogens who presented with abdominal pain for two months. Diagnostic work-up revealed elevated gamma-glutamyl transferase and lactate dehydrogenase levels. Abdominal ultrasound described a large nodular lesion in the right lobe of the liver described as a hemangioma. One month later, a computed tomography-scan was made and revealed the same lesion, which had grown from 13.5 to 20 cm, maintaining typical imaging characteristics of a hemangioma. A right hepatectomy was performed and pathology revealed an angiosarcoma. After surgery, a positron emission tomography-com- puted tomography scan showed hepatic and bone metastasis. The patient started taxane-based chemotherapy and lumbar palliative radiotherapy, but died 10 months after surgery. This case shows how difficult it is to diagnose hepatic angiosarcoma relying only on imaging findings. Two abdominal computed tomography -scans were performed and none suggested this diagnosis. Angiosarcoma is a very aggressive tumour with an adverse prognosis. Surgery is the only curative treatment available. However, it is rarely feasible due to unresectable disease or distant metastasis. -

Hepatic Angiosarcoma: Mimicking of Angioma on Three-Phase Technetium-99M Red Blood Cell Scintigraphy
Hepatic Angiosarcoma: Mimicking of Angioma on Three-Phase Technetium-99m Red Blood Cell Scintigraphy Ferris Ginsberg, James D. Slavin, Jr., and Richard P. Spencer Department of Radiology, Saint Francis Hospital and Medical Center, Hartford; and Department of Nuclear Medicine, University of Connecticut Health Center, Farmington, Connecticut A [99mTc]RBC study in a 63-yr-old man showed intrahepatic lesions which initially had less activity than surrounding liver tissue. When viewed 3 hr later, these had "reversed" and the lesions revealed increased uptake of the radiolabeled red cells. Some extrahepatic areas showed the same pattern (these were in the mesentery of the small bowel). The lesions proved to be angiosarcomas. Hence, the behavior of labeled red cells in these angiosarcomas mimicked that in benign hemangiomas. J NucÃMed 27:1861-1863,1986 . he three-phase technetium-99m red cell ([WmTc] ~ 1,500 rad external radiation to the region. During a chole- RBC) imaging technique has been reported as highly cystectomy performed 10 yr prior to the current admission, specific for identifying benign intrahepatic heman careful exploration of the abdomen revealed only thickening giomas; the imaging pattern has not been described in of the left retroperitoneum. There was no evidence of tumor any other lesion (1-3). We describe a case with an at that time and the liver was noted to be normal. imaging study identical to that of intrahepatic heman An exploratory laparotomy was performed 3 wk after ad mission; it revealed multiple intrahepatic masses as well as giomas, but which was due to a malignant tumor. multiple smaller nodular lesions involving most of the small bowel mesentery. -

Angiosarcoma and Hemangioendothelioma
ANGIOSARCOMA AND HEMANGIOENDOTHELIOMA Claudia Mª Valverde Vall d´Hebrón University Hospital VASCULAR TUMORS BENIGN-> - Hemangioma BORDERLINE - Hemangioendothelioma MALIGN-> - AiAngiosarcoma -Kaposi VASCULAR TUMORS BENIGN-> - Hemangioma BORDERLINE - Hemangioendothelioma MALIGN-> - AiAngiosarcoma -Kaposi HEMANGIOENDOTHELIOMA( HE) Vascular tumors with a biologic behaviour intermediate between hemangioma and angiosarcoma: Ability to recur locally and some to metastatize but at a far reduced rate comppgared with angiosarcoma Subtypes: - Epithelioid HE - Kaposiform HE - Hobnail HE - Epithelioid sarcoma-like HE EPITHELIOID EH Clinical features: - Rare in childhood - Both sexes equally - Usually solitary, slightly painful mass SOFT TISSUE BONE LIVER Weiss et al Kleer et al Makhlouf et al Local recurrence 13% Metastasis 31% 31% 61% Mortality 13% 31% 43% Weiss et al. Semin diagn pathol 1986. Kleer et al. Am J Surg Pathol 1996. Makhlouf et al . Cancer 1999. EPITHELIOID EH • Pathological features - Angiocentric - Vascular differentiation more primitive - Short strands or solid nests of rounded/slightly spindled endothelial cells. - Form small intracellular lumens –”vacuoles” - Atypia, >1mit/10HPF, spindling or necrosis-> more agressive Weiss et al. Semin diagn pathol 1986 KAPOSIFORM HE Clinical Features: - Childhoo d - Trunk, retoperitoneum. - Kasabach-Merritt phenomenon - ill defined violaceus ppqlaque - No tendency to regress - 10% mortality - Virtually no metastatic. KAPOSIFORM HE • Pathological features: • Small CD31+ vessels surrounded by actin- -
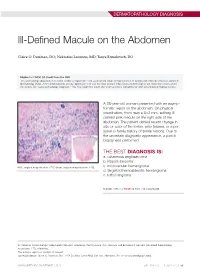
Ill-Defined Macule on the Abdomen
DERMATOPATHOLOGY DIAGNOSIS Ill-Defined Macule on the Abdomen Claire O. Dorfman, DO; Nektarios Lountzis, MD; Tanya Ermolovich, DO Eligible for 1 MOC SA Credit From the ABD This Dermatology Diagnosis in our print edition is eligible for 1 self-assessment credit for Maintenance of Certification from the American Board of Dermatology (ABD). After completing this activity, diplomates can visit the ABD website (http://www.abderm.org) to self-report the credits under the activity title “Cutis Dermatology Diagnosis.” You may report the credit after each activity is completed or after accumulating multiple credits. A 38-year-old woman presented with an asymp- tomatic lesion on the abdomen. On physical examination, there was a 5×2-mm, solitary, ill- defined pink macule on the right side of the abdomen. The patient denied recent change in size or color of the lesion, prior trauma, or a per- sonal or family history of similar lesions. Due to the uncertain diagnostic appearance, a punch biopsy was performed. THE BEST DIAGNOSIS IS: a. cutaneous angiosarcoma b. Kaposi sarcoma H&E, original magnification ×200 (inset, original magnification ×40). c. microvenular hemangioma d. targetoid hemosiderotic hemangioma e. tufted angioma PLEASE TURN TO PAGE 68 FOR THE DIAGNOSIS Dr. Dorfman is from Lehigh Valley Health Network, Allentown, Pennsylvania. Drs. Lountzis and Ermolovich are from Advanced Dermatology Associates, LTD, Allentown. The authors report no conflict of interest. Correspondence: Claire O. Dorfman, DO, 1259 S Cedar Crest Blvd, Ste 100, Allentown, PA 18103 ([email protected]). WWW.MDEDGE.COM/DERMATOLOGY VOL. 104 NO. 1 I JULY 2019 53 DERMATOPATHOLOGY DIAGNOSIS DISCUSSION THE DIAGNOSIS: Microvenular Hemangioma icrovenular hemangioma is an acquired benign vas- from pink patches to dark purple plaques or nodules that cular neoplasm that was described by Hunt et al1 in commonly occur on the lower legs10; however, the clinical M1991, though Bantel et al2 reported a similar entity appearance of KS varies depending on the clinical variant termed micropapillary angioma in 1989. -

Purpuric Macule of the Right Axilla
DERMATOPATHOLOGY DIAGNOSIS Purpuric Macule of the Right Axilla Andrew L.J. Dunn, MD; Brett H. Keeling, MD; Justin P. Bandino, MD; Dirk M. Elston, MD; John S. Metcalf, MD Eligible for 1 MOC SA Credit From the ABD This Dermatopathology Diagnosis article in our print edition is eligible for 1 self-assessment credit for Maintenance of Certification from the American Board of Dermatology (ABD). After completing this activity, diplomates can visit the ABD website (http://www.abderm.org) to self-report the credits under the activity title “Cutis Dermatopathology Diagnosis.” You may report the credit after each activity is completed or after accumu- lating multiple credits. A 67-year-old woman presented with a lesion on the medial aspect of the right axilla of 2 weeks’ duration. The patientcopy had a history of cancer of the right breast treated with a mastectomy and adjuvant radiation. She denied pain, bleeding, pruritus, or rapid growth, as well as any changes in medicationnot or recent trauma. Physical examina- tion revealed a 5-mm purpuric macule of the right axilla. A punch biopsy was performed. Amplifica- tion for the C-MYC gene was negative by fluores- Docence in situ hybridization. THE BEST DIAGNOSIS IS: a. atypical vascular lesion H&E, original magnification ×100 (Ki-67 immunostain, original magnifi- cation ×100 [inset]). b. hobnail hemangioma (targetoid hemosiderotic hemangioma) c. Kaposi sarcoma CUTIS d. lymphangioma circumscriptum (superficial lymphangioma) e. radiation-induced angiosarcoma PLEASE TURN TO PAGE 29 FOR THE DIAGNOSIS Dr. Dunn is from the Department of Pathology and Laboratory Services, University of Arkansas for Medical Sciences, Little Rock. -

Angiosarcoma: a Case Report and Review of the Literature
CONTINUING MEDICAL EDUCATION Angiosarcoma: A Case Report and Review of the Literature Abdulhafez Selim, MD, PhD; Amor Khachemoune, MD, CWS; Norman A. Lockshin, MD GOAL To understand angiosarcoma to better manage patients with the condition OBJECTIVES Upon completion of this activity, dermatologists and general practitioners should be able to: 1. Describe the clinical presentation of angiosarcoma. 2. Identify the differential diagnoses of angiosarcoma. 3. Discuss the treatment options for patients with angiosarcoma. CME Test on page 309. This article has been peer reviewed and is accredited by the ACCME to provide continuing approved by Michael Fisher, MD, Professor of medical education for physicians. Medicine, Albert Einstein College of Medicine. Albert Einstein College of Medicine designates Review date: October 2005. this educational activity for a maximum of 1 This activity has been planned and implemented category 1 credit toward the AMA Physician’s in accordance with the Essential Areas and Policies Recognition Award. Each physician should of the Accreditation Council for Continuing Medical claim only that credit that he/she actually spent Education through the joint sponsorship of Albert in the activity. Einstein College of Medicine and Quadrant This activity has been planned and produced in HealthCom, Inc. Albert Einstein College of Medicine accordance with ACCME Essentials. Drs. Selim, Khachemoune, and Lockshin report no conflict of interest. The authors report no discussion of off-label use. Dr. Fisher reports no conflict of interest. Angiosarcoma is an aggressive neoplasm that includes a multidisciplinary team and may involve predominantly affects elderly patients. Most a combination of surgery, radiation, and chemo- cases appear on the scalp and face de novo; therapy tailored to the patient’s age and associ- however, trauma, longstanding lymphedema, and ated comorbidities. -

Targetoid Hemosiderotic Hemangioma (Hobnail Hemangioma): a Case Report
12 Scientific Journal of Kurdistan University of Medical Sciences No.92/Feb-Mar 2018 Targetoid hemosiderotic hemangioma (hobnail hemangioma): A case report Shamsi Meymandi S., MD1, Khalili M., MD2, Aflatoonian M., MD3 1. Professor, Dermatology Department, Pathology and Stem Cell Research Center, Kerman University of Medical Sciences, Kerman, Iran. 2. Assistant Professor, Dermatology Department, Kerman University of Medical Sciences, Kerman, Iran. 3. Assistant Professor, Dermatology Department, Kerman University of Medical Sciences, Kerman, Iran (Corresponding Author), Iran. Tel:+98-34-31328328, [email protected] ABSTRACT Background and Aim: Hobnail hemangioma ( targetoid hemosiderotic hemangioma) is a benign vascular tumor that is usually characterized as a papule with an ecchymotic halo on the periphery. It is usually seen in young adults and most commonly affects limbs. The lesion is seen equally in both sexes. This vascular tumor is rare in the world and, according to our acknowledge, only one case has been reported in Iran. In this case report, we described the second case of targetoid hemosiderotic hemangioma. Material and Methods: The patient is an 8 years old boy with a targetoid asymptomatic red popular lesion on his lower leg from 5 years ago .The lesion was completely excised. Skin biopsy showed dilated vascular spaces with hobnail endothelial cells in the superficial dermis accompanied with narrower vessels in the deep dermis which was compatible with targetoid hemosiderotic hemangioma. In a follow up period of three years the lesion did not recur. Conclusion: Distinction between targetoid hemosiderotic hemangioma and malignant skin lesions such as melanoma, Kaposi sarcoma and angiosarcoma is necessary. Thus, knowledge of clinical picture and pathology of this lesion can prevent unnecessary aggressive procedures. -

Abstract Hobnail Hemangioma Is a Rare, Benign Vascular Growth That Typically Presents in the Third and Fourth Decades of Life
Volume 19 Number 5 May 2013 Photo Vignette Hobnail Hemangioma on the Trunk C.Vijay Krishna MD, G. Madhusudhan Reddy MBBS, Senthil Kumar A.L. MBBS, A.Vijaya Mohan Rao MD Dermatology Online Journal 19 (5): 10 Department of Dermatology, Venereology, and Leprology, Narayana Medical College & Hospital, Nellore-524002, India Correspondence: C. Vijay Krishna, MD Assistant Professor, Department of Dermatology, Narayana Medical College & Hospital, Nellore-524002, India E-mail: [email protected] Phone No: 91-9491042940 Fax: 91- 0861 - 2331763 Abstract Hobnail hemangioma is a rare, benign vascular growth that typically presents in the third and fourth decades of life. It classically presents as a targetoid lesion with a violaceous central papule surrounded by a peripheral ecchymotic rim. Common sites of involvement include extremities and trunk. We present this case of hobnail hemangioma in a 10-year- old boy because of its rarity. Keywords: Hobnail Hemangioma Figure 1. A well-defined plaque on the trunk with a violaceous centre and ecchymotic rim giving a “targetoid” appearance Case synopsis A 10-year-old boy presented with a 7-year history of a single, asymptomatic plaque on the right side of the trunk (Fig. 1). The lesion started as an erythematous papule and later gradually increased in size. Cutaneous examination revealed a single non-tender oval plaque of 2x3 cm, present on the right side of the trunk. The plaque had a violaceous center and a peripheral ecchymotic rim, giving a targetoid appearance. The remainder of the skin examination was unremarkable. There was no history of fluid filled lesions, trauma, or insect bite preceding the lesion.