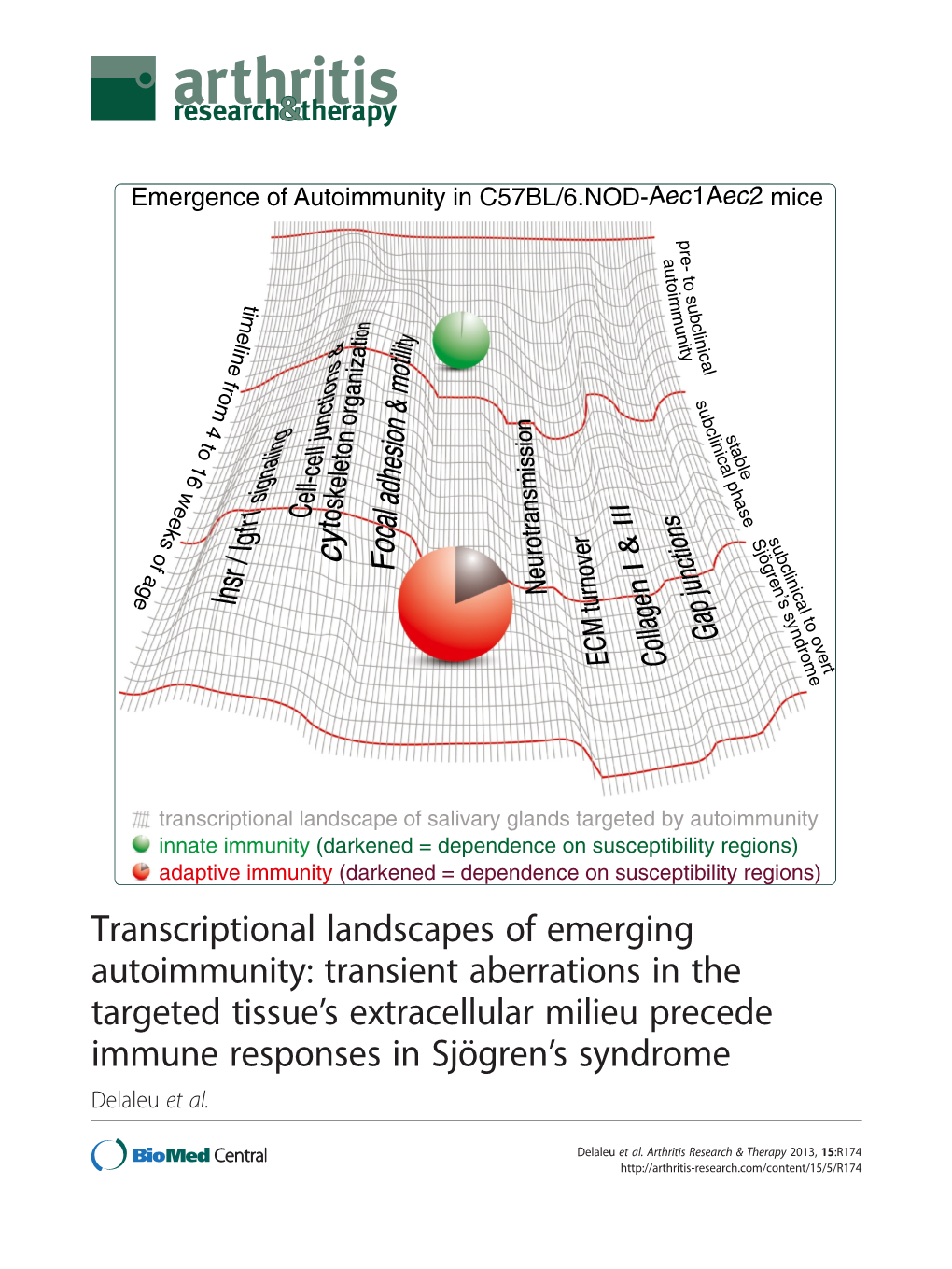
Transcriptional Landscapes of Emerging Autoimmunity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Release Β ATP-Induced IL-1 and Canonical Nicotinic Agonists Inhibit Phosphocholine-Modified Macromolecules
Phosphocholine-Modified Macromolecules and Canonical Nicotinic Agonists Inhibit ATP-Induced IL-1β Release This information is current as Andreas Hecker, Mira Küllmar, Sigrid Wilker, Katrin of September 24, 2021. Richter, Anna Zakrzewicz, Srebrena Atanasova, Verena Mathes, Thomas Timm, Sabrina Lerner, Jochen Klein, Andreas Kaufmann, Stefan Bauer, Winfried Padberg, Wolfgang Kummer, Sabina Janciauskiene, Martin Fronius, Elke K. H. Schweda, Günter Lochnit and Veronika Grau Downloaded from J Immunol 2015; 195:2325-2334; Prepublished online 22 July 2015; doi: 10.4049/jimmunol.1400974 http://www.jimmunol.org/content/195/5/2325 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2015/07/22/jimmunol.140097 Material 4.DCSupplemental References This article cites 42 articles, 11 of which you can access for free at: http://www.jimmunol.org/content/195/5/2325.full#ref-list-1 by guest on September 24, 2021 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2015 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. -

Global Analysis Reveals the Complexity of the Human Glomerular Extracellular Matrix
Global analysis reveals the complexity of the human glomerular extracellular matrix Rachel Lennon,1,2 Adam Byron,1,* Jonathan D. Humphries,1 Michael J. Randles,1,2 Alex Carisey,1 Stephanie Murphy,1,2 David Knight,3 Paul E. Brenchley,2 Roy Zent,4,5 and Martin J. Humphries.1 1Wellcome Trust Centre for Cell-Matrix Research, Faculty of Life Sciences, University of Manchester, Manchester, UK; 2Faculty of Medical and Human Sciences, University of Manchester, Manchester, UK; 3Biological Mass Spectrometry Core Facility, Faculty of Life Sciences, University of Manchester, Manchester, UK; 4Division of Nephrology, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA; and 5Veterans Affairs Hospital, Nashville, TN, USA. *Present address: Edinburgh Cancer Research UK Centre, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK. Running title: Proteome of the glomerular matrix Word count: Abstract: 208, main text 2765 Corresponding author: Dr Rachel Lennon, Wellcome Trust Centre for Cell-Matrix Research, Michael Smith Building, University of Manchester, Manchester M13 9PT, UK. Phone: 0044 (0) 161 2755498. Fax: 0044 (0) 161 2755082. Email: [email protected] Abstract The glomerulus contains unique cellular and extracellular matrix (ECM) components, which are required for intact barrier function. Studies of the cellular components have helped to build understanding of glomerular disease; however, the full composition and regulation of glomerular ECM remains poorly understood. Here, we employed mass spectrometry–based proteomics of enriched ECM extracts for a global analysis of human glomerular ECM in vivo and identified a tissue-specific proteome of 144 structural and regulatory ECM proteins. This catalogue includes all previously identified glomerular components, plus many new and abundant components. -

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Environmental Influences on Endothelial Gene Expression
ENDOTHELIAL CELL GENE EXPRESSION John Matthew Jeff Herbert Supervisors: Prof. Roy Bicknell and Dr. Victoria Heath PhD thesis University of Birmingham August 2012 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. ABSTRACT Tumour angiogenesis is a vital process in the pathology of tumour development and metastasis. Targeting markers of tumour endothelium provide a means of targeted destruction of a tumours oxygen and nutrient supply via destruction of tumour vasculature, which in turn ultimately leads to beneficial consequences to patients. Although current anti -angiogenic and vascular targeting strategies help patients, more potently in combination with chemo therapy, there is still a need for more tumour endothelial marker discoveries as current treatments have cardiovascular and other side effects. For the first time, the analyses of in-vivo biotinylation of an embryonic system is performed to obtain putative vascular targets. Also for the first time, deep sequencing is applied to freshly isolated tumour and normal endothelial cells from lung, colon and bladder tissues for the identification of pan-vascular-targets. Integration of the proteomic, deep sequencing, public cDNA libraries and microarrays, delivers 5,892 putative vascular targets to the science community. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Protein Identities in Evs Isolated from U87-MG GBM Cells As Determined by NG LC-MS/MS
Protein identities in EVs isolated from U87-MG GBM cells as determined by NG LC-MS/MS. No. Accession Description Σ Coverage Σ# Proteins Σ# Unique Peptides Σ# Peptides Σ# PSMs # AAs MW [kDa] calc. pI 1 A8MS94 Putative golgin subfamily A member 2-like protein 5 OS=Homo sapiens PE=5 SV=2 - [GG2L5_HUMAN] 100 1 1 7 88 110 12,03704523 5,681152344 2 P60660 Myosin light polypeptide 6 OS=Homo sapiens GN=MYL6 PE=1 SV=2 - [MYL6_HUMAN] 100 3 5 17 173 151 16,91913397 4,652832031 3 Q6ZYL4 General transcription factor IIH subunit 5 OS=Homo sapiens GN=GTF2H5 PE=1 SV=1 - [TF2H5_HUMAN] 98,59 1 1 4 13 71 8,048185945 4,652832031 4 P60709 Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 - [ACTB_HUMAN] 97,6 5 5 35 917 375 41,70973209 5,478027344 5 P13489 Ribonuclease inhibitor OS=Homo sapiens GN=RNH1 PE=1 SV=2 - [RINI_HUMAN] 96,75 1 12 37 173 461 49,94108966 4,817871094 6 P09382 Galectin-1 OS=Homo sapiens GN=LGALS1 PE=1 SV=2 - [LEG1_HUMAN] 96,3 1 7 14 283 135 14,70620005 5,503417969 7 P60174 Triosephosphate isomerase OS=Homo sapiens GN=TPI1 PE=1 SV=3 - [TPIS_HUMAN] 95,1 3 16 25 375 286 30,77169764 5,922363281 8 P04406 Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 - [G3P_HUMAN] 94,63 2 13 31 509 335 36,03039959 8,455566406 9 Q15185 Prostaglandin E synthase 3 OS=Homo sapiens GN=PTGES3 PE=1 SV=1 - [TEBP_HUMAN] 93,13 1 5 12 74 160 18,68541938 4,538574219 10 P09417 Dihydropteridine reductase OS=Homo sapiens GN=QDPR PE=1 SV=2 - [DHPR_HUMAN] 93,03 1 1 17 69 244 25,77302971 7,371582031 11 P01911 HLA class II histocompatibility antigen, -

Original Article FREM2 Is an Independent Predictor of Poor Survival in Clear Cell Renal Cell Carcinoma-Evidence from the Cancer Genome Atlas (TCGA)
Int J Clin Exp Med 2019;12(12):13741-13748 www.ijcem.com /ISSN:1940-5901/IJCEM0076963 Original Article FREM2 is an independent predictor of poor survival in clear cell renal cell carcinoma-evidence from the cancer genome atlas (TCGA) Weiping Huang, Yongyong Lu, Xixi Huang, Feng Wang, Zhixian Yu Department of Urology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325035, Zhejiang Province, China Received February 26, 2018; Accepted October 7, 2018; Epub December 15, 2019; Published December 30, 2019 Abstract: Fraser syndrome protein 1 (FRAS1) and FRAS1 related extracellular matrix protein 1 and 2 (FREM1, FREM2) are a novel group of basement membrane proteins. The relationship between the three gene (FRAS1, FREM1, FREM2) and renal clear cell carcinoma is completely unclear. Thus, in this research, we used the mRNA sequencing data derived from TCGA kidney renal clear cell carcinoma cohort to assess the association of FRAS1, FREM1 and FREM2 with different clinical features. FRAS1, FREM1 and FREM2 mRNA levels were downregulated in KIRC (kidney renal clear cell carcinoma) tissues than normal tissues (FRAS1, P < 0.0001; FREM1, P < 0.0001, FREM2, P = 0.0001), respectively. FRAS1, FREM1 and FREM2 were significantly different in histologic grade, patho- logic stage and pathologic T (all P < 0.001). Low FRAS1, FREM1 and FREM2 expression were correlated to worsen overall survival (all P < 0.01), and Low FREM1 and FREM2 expression had worse relapse-free survival (FREM1, P = 0.0113; FREM2, P = 0.0424). Multivariate Cox regression analysis revealed that FREM2 was an independent prog- nostic factor for overall survival. Taken together, FREM2 expression is an independent predictor of poor survival in renal clear cell carcinoma and is positively associated with advanced stage, high histologic grade. -

Download Download
Supplementary Figure S1. Results of flow cytometry analysis, performed to estimate CD34 positivity, after immunomagnetic separation in two different experiments. As monoclonal antibody for labeling the sample, the fluorescein isothiocyanate (FITC)- conjugated mouse anti-human CD34 MoAb (Mylteni) was used. Briefly, cell samples were incubated in the presence of the indicated MoAbs, at the proper dilution, in PBS containing 5% FCS and 1% Fc receptor (FcR) blocking reagent (Miltenyi) for 30 min at 4 C. Cells were then washed twice, resuspended with PBS and analyzed by a Coulter Epics XL (Coulter Electronics Inc., Hialeah, FL, USA) flow cytometer. only use Non-commercial 1 Supplementary Table S1. Complete list of the datasets used in this study and their sources. GEO Total samples Geo selected GEO accession of used Platform Reference series in series samples samples GSM142565 GSM142566 GSM142567 GSM142568 GSE6146 HG-U133A 14 8 - GSM142569 GSM142571 GSM142572 GSM142574 GSM51391 GSM51392 GSE2666 HG-U133A 36 4 1 GSM51393 GSM51394 only GSM321583 GSE12803 HG-U133A 20 3 GSM321584 2 GSM321585 use Promyelocytes_1 Promyelocytes_2 Promyelocytes_3 Promyelocytes_4 HG-U133A 8 8 3 GSE64282 Promyelocytes_5 Promyelocytes_6 Promyelocytes_7 Promyelocytes_8 Non-commercial 2 Supplementary Table S2. Chromosomal regions up-regulated in CD34+ samples as identified by the LAP procedure with the two-class statistics coded in the PREDA R package and an FDR threshold of 0.5. Functional enrichment analysis has been performed using DAVID (http://david.abcc.ncifcrf.gov/) -

Understanding the Role of the Chromosome 15Q25.1 in COPD Through Epigenetics and Transcriptomics
European Journal of Human Genetics (2018) 26:709–722 https://doi.org/10.1038/s41431-017-0089-8 ARTICLE Understanding the role of the chromosome 15q25.1 in COPD through epigenetics and transcriptomics 1 1,2 1,3,4 5,6 5,6 Ivana Nedeljkovic ● Elena Carnero-Montoro ● Lies Lahousse ● Diana A. van der Plaat ● Kim de Jong ● 5,6 5,7 6 8 9 10 Judith M. Vonk ● Cleo C. van Diemen ● Alen Faiz ● Maarten van den Berge ● Ma’en Obeidat ● Yohan Bossé ● 11 1,12 12 1 David C. Nickle ● BIOS Consortium ● Andre G. Uitterlinden ● Joyce J. B. van Meurs ● Bruno C. H. Stricker ● 1,4,13 6,8 5,6 1 1 Guy G. Brusselle ● Dirkje S. Postma ● H. Marike Boezen ● Cornelia M. van Duijn ● Najaf Amin Received: 14 March 2017 / Revised: 6 November 2017 / Accepted: 19 December 2017 / Published online: 8 February 2018 © The Author(s) 2018. This article is published with open access Abstract Chronic obstructive pulmonary disease (COPD) is a major health burden in adults and cigarette smoking is considered the most important environmental risk factor of COPD. Chromosome 15q25.1 locus is associated with both COPD and smoking. Our study aims at understanding the mechanism underlying the association of chromosome 15q25.1 with COPD through epigenetic and transcriptional variation in a population-based setting. To assess if COPD-associated variants in 1234567890();,: 15q25.1 are methylation quantitative trait loci, epigenome-wide association analysis of four genetic variants, previously associated with COPD (P < 5 × 10−8) in the 15q25.1 locus (rs12914385:C>T-CHRNA3, rs8034191:T>C-HYKK, rs13180: C>T-IREB2 and rs8042238:C>T-IREB2), was performed in the Rotterdam study (n = 1489). -

Scrutiny of the CHRNA5-CHRNA3-CHRNB4
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2013 Scrutiny of the CHRNA5-CHRNA3-CHRNB4 smoking behavior locus reveals a novel association with alcohol use in a Finnish population based study Jenni Hällfors University of Helsinki Anu Loukola University of Helsinki Janne Pitkäniemi University of Helsinki Ulla Broms University of Helsinki Satu Männistö National Institute for Health and Welfare, Helsinki, Finland See next page for additional authors Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Recommended Citation Hällfors, Jenni; Loukola, Anu; Pitkäniemi, Janne; Broms, Ulla; Männistö, Satu; Salomaa, Veikko; Heliövaara, Markku; Lehtimäki, Terho; Raitakari, Olli; Madden, Pamela AF; Heath, Andrew C.; Montgomery, Grant W.; Martin, Nicholas G.; Korhonen, Tellervo; and Kaprio, Jaakko, ,"Scrutiny of the CHRNA5-CHRNA3-CHRNB4 smoking behavior locus reveals a novel association with alcohol use in a Finnish population based study." International Journal of Molecular Epidemiology and Genetics.4,2. 109-119. (2013). https://digitalcommons.wustl.edu/open_access_pubs/3075 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Jenni Hällfors, Anu Loukola, Janne Pitkäniemi, Ulla Broms, Satu Männistö, Veikko Salomaa, Markku Heliövaara, Terho Lehtimäki, -

Gene Expression Studies in Depression Development and Treatment
Mariani et al. Translational Psychiatry (2021) 11:354 https://doi.org/10.1038/s41398-021-01469-6 Translational Psychiatry REVIEW ARTICLE Open Access Gene expression studies in Depression development and treatment: an overview of the underlying molecular mechanisms and biological processes to identify biomarkers Nicole Mariani 1, Nadia Cattane2,CarminePariante 1 and Annamaria Cattaneo 2,3 Abstract A combination of different risk factors, such as genetic, environmental and psychological factors, together with immune system, stress response, brain neuroplasticity and the regulation of neurotransmitters, is thought to lead to the development of major depressive disorder (MDD). A growing number of studies have tried to investigate the underlying mechanisms of MDD by analysing the expression levels of genes involved in such biological processes. These studies have shown that MDD is not just a brain disorder, but also a body disorder, and this is mainly due to the interplay between the periphery and the Central Nervous System (CNS). To this purpose, most of the studies conducted so far have mainly dedicated to the analysis of the gene expression levels using postmortem brain tissue as well as peripheral blood samples of MDD patients. In this paper, we reviewed the current literature on candidate gene expression alterations and the few existing transcriptomics studies in MDD focusing on inflammation, neuroplasticity, neurotransmitters and stress-related genes. Moreover, we focused our attention on studies, which have investigated 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; mRNA levels as biomarkers to predict therapy outcomes. This is important as many patients do not respond to antidepressant medication or could experience adverse side effects, leading to the interruption of treatment. -

Research Article Microarray-Based Comparisons of Ion Channel Expression Patterns: Human Keratinocytes to Reprogrammed Hipscs To
Hindawi Publishing Corporation Stem Cells International Volume 2013, Article ID 784629, 25 pages http://dx.doi.org/10.1155/2013/784629 Research Article Microarray-Based Comparisons of Ion Channel Expression Patterns: Human Keratinocytes to Reprogrammed hiPSCs to Differentiated Neuronal and Cardiac Progeny Leonhard Linta,1 Marianne Stockmann,1 Qiong Lin,2 André Lechel,3 Christian Proepper,1 Tobias M. Boeckers,1 Alexander Kleger,3 and Stefan Liebau1 1 InstituteforAnatomyCellBiology,UlmUniversity,Albert-EinsteinAllee11,89081Ulm,Germany 2 Institute for Biomedical Engineering, Department of Cell Biology, RWTH Aachen, Pauwelstrasse 30, 52074 Aachen, Germany 3 Department of Internal Medicine I, Ulm University, Albert-Einstein Allee 11, 89081 Ulm, Germany Correspondence should be addressed to Alexander Kleger; [email protected] and Stefan Liebau; [email protected] Received 31 January 2013; Accepted 6 March 2013 Academic Editor: Michael Levin Copyright © 2013 Leonhard Linta et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Ion channels are involved in a large variety of cellular processes including stem cell differentiation. Numerous families of ion channels are present in the organism which can be distinguished by means of, for example, ion selectivity, gating mechanism, composition, or cell biological function. To characterize the distinct expression of this group of ion channels we have compared the mRNA expression levels of ion channel genes between human keratinocyte-derived induced pluripotent stem cells (hiPSCs) and their somatic cell source, keratinocytes from plucked human hair. This comparison revealed that 26% of the analyzed probes showed an upregulation of ion channels in hiPSCs while just 6% were downregulated.