Towards Automated Molecular Search in Drug Space Rustam Zhumagambetov
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

The Promise of Human Genome Editing for Rare and Genetic Disease Summary Report of the 2019 FORUM Annual Lecture
The promise of human genome editing for rare and genetic disease Summary report of the 2019 FORUM Annual Lecture The Academy of Medical Sciences The Academy of Medical Sciences is the independent body in the UK representing the diversity of medical science. Our mission is to promote medical science and its translation into benefits for society. The Academy’s elected Fellows are the United Kingdom’s leading medical scientists from hospitals, academia, industry and the public service. We work with them to promote excellence, influence policy to improve health and wealth, nurture the next generation of medical researchers, link academia, industry and the NHS, seize international opportunities and encourage dialogue about the medical sciences. Opinions expressed in this report do not necessarily represent the views of all participants at the event, the Academy of Medical Sciences, or its Fellows. All web references were accessed in February 2020. This work is © Academy of Medical Sciences and is licensed under Creative Commons Attribution 4.0 International. The Academy of Medical Sciences 3 The promise of human genome editing for rare and genetic disease Summary report of the 2019 FORUM Annual Lecture Contents Executive summary .................................................................................................... 4 An introduction to genome editing ................................................................................ 6 Genome editing: moving to the clinic ........................................................................... -

BBSRC Annual Report and Accounts 2007-08 HC
ANNUAL REPORT & ACCOUNTS 2007-2008 Biotechnology and Biological Sciences Research Council ANNUAL REPORT & ACCOUNTS 2007-2008 Presented to Parliament by the Secretary of State, and by the Comptroller and Auditor General, in pursuance of Schedule 1, Sections 2 [2] and 3 [3] of the Science and Technology Act 1965. Ordered to be printed by the House of Commons 16 July 2008. HC761 LONDON: The Stationery Office £18.55 Contents PART 1: MANAGEMENT COMMENTARY Chairman’s statement 1 Chief Executive’s report 3 Supporting world class research 6 Key funding data 7 Embedding Systems Biology 11 Major collaborative and multidisciplinary programmes 12 Delivering economic and social benefits 14 Collaborative research with industry 14 Commercialising research outputs 17 Tackling major challenges 20 People, skills, training and knowledge flows 22 Embedding our science in society 26 Opinion gathering and public dialogue 26 Outreach and engagement 28 Engaging young people in science 29 Corporate information 30 2007-2008 Council 30 Boards, Panels and Committees 31 Organisational developments 36 Financial review 39 Remuneration report 42 PART 2: ANNUAL ACCOUNTS Financial statements for the year ended 31 March 2008 45 This Annual Report covers the period from 1 April 2007 to 31 March 2008. BBSRC ANNUAL REPORT & ACCOUNTS BBSRC ANNUAL The Biotechnology and Biological Sciences Research Council (BBSRC), established by Royal Charter in 1994, is the UK’s principal funder of basic and strategic research across the biosciences (www. bbsrc.ac.uk). It is funded primarily by the Science Budget, through the Department for Innovation, Universities and Skills (DIUS). Our mission is to support high-class science and research training, and to promote knowledge transfer in support of bio-based industries and public engagement in bioscience. -

Winter for the Membership of the American Crystallographic Association, P.O
AMERICAN CRYSTALLOGRAPHIC ASSOCIATION NEWSLETTER Number 4 Winter 2004 ACA 2005 Transactions Symposium New Horizons in Structure Based Drug Discovery Table of Contents / President's Column Winter 2004 Table of Contents President's Column Presidentʼs Column ........................................................... 1-2 The fall ACA Council Guest Editoral: .................................................................2-3 meeting took place in early 2004 ACA Election Results ................................................ 4 November. At this time, News from Canada / Position Available .............................. 6 Council made a few deci- sions, based upon input ACA Committee Report / Web Watch ................................ 8 from the membership. First ACA 2004 Chicago .............................................9-29, 38-40 and foremost, many will Workshop Reports ...................................................... 9-12 be pleased to know that a Travel Award Winners / Commercial Exhibitors ...... 14-23 satisfactory venue for the McPherson Fankuchen Address ................................38-40 2006 summer meeting was News of Crystallographers ...........................................30-37 found. The meeting will be Awards: Janssen/Aminoff/Perutz ..............................30-33 held at the Sheraton Waikiki Obituaries: Blow/Alexander/McMurdie .................... 33-37 Hotel in Honolulu, July 22-27, 2005. Council is ACA Summer Schools / 2005 Etter Award ..................42-44 particularly appreciative of Database Update: -

Astex Pharmaceuticals Announces the Appointment of President Harren Jhoti, Phd to the Bioindustry Association Board of Directors
October 10, 2012 Astex Pharmaceuticals Announces the Appointment of President Harren Jhoti, PhD to the BioIndustry Association Board of Directors DUBLIN, Calif., Oct. 10, 2012 (GLOBE NEWSWIRE) -- Astex Pharmaceuticals, Inc. (Nasdaq:ASTX), a pharmaceutical company dedicated to the discovery and development of novel small molecule therapeutics, announced that Harren Jhoti, PhD, president and director of Astex Pharmaceuticals, was appointed to the BioIndustry Association (BIA) Board of Directors. About Harren Jhoti, PhD Dr. Harren Jhoti has served as Astex Pharmaceuticals president and member of the Board of Directors since the company's formation in July 2011. He co-founded Astex Therapeutics in 1999 and was chief scientific officer until November 2007 when he was appointed chief executive. Dr. Jhoti was awarded the Prous Institute-Overton and Meyer Award for New Technologies in Drug Discovery by the European Federation for Medicinal Chemistry in 2012 and was also named by the Royal Society of Chemistry as "Chemistry World Entrepreneur of the Year" for 2007. He has published widely including in leading journals such as Nature and Science and has also been featured in TIME magazine after being named by the World Economic Forum a Technology Pioneer in 2005. Dr. Jhoti served as a non-executive director of Iconix Inc. Before starting up Astex Therapeutics in 1999, he was head of Structural Biology and Bioinformatics at GlaxoWellcome in the United Kingdom (1991-1999). Prior to Glaxo, Dr. Jhoti was a post-doctoral scientist at Oxford University. He received a BSc (Hons) in Biochemistry in 1985 and a PhD in Protein Crystallography from the University of London in 1989. -

Anew Drug Design Strategy in the Liht of Molecular Hybridization Concept
www.ijcrt.org © 2020 IJCRT | Volume 8, Issue 12 December 2020 | ISSN: 2320-2882 “Drug Design strategy and chemical process maximization in the light of Molecular Hybridization Concept.” Subhasis Basu, Ph D Registration No: VB 1198 of 2018-2019. Department Of Chemistry, Visva-Bharati University A Draft Thesis is submitted for the partial fulfilment of PhD in Chemistry Thesis/Degree proceeding. DECLARATION I Certify that a. The Work contained in this thesis is original and has been done by me under the guidance of my supervisor. b. The work has not been submitted to any other Institute for any degree or diploma. c. I have followed the guidelines provided by the Institute in preparing the thesis. d. I have conformed to the norms and guidelines given in the Ethical Code of Conduct of the Institute. e. Whenever I have used materials (data, theoretical analysis, figures and text) from other sources, I have given due credit to them by citing them in the text of the thesis and giving their details in the references. Further, I have taken permission from the copyright owners of the sources, whenever necessary. IJCRT2012039 International Journal of Creative Research Thoughts (IJCRT) www.ijcrt.org 284 www.ijcrt.org © 2020 IJCRT | Volume 8, Issue 12 December 2020 | ISSN: 2320-2882 f. Whenever I have quoted written materials from other sources I have put them under quotation marks and given due credit to the sources by citing them and giving required details in the references. (Subhasis Basu) ACKNOWLEDGEMENT This preface is to extend an appreciation to all those individuals who with their generous co- operation guided us in every aspect to make this design and drawing successful. -

Bioinformatics
Bioinformatics Antal, Péter Hullám, Gábor Millinghoffer, András Hajós, Gergely Marx, Péter Arany, Ádám Bolgár, Bence Gézsi, András Sárközy, Péter Poppe, László Created by XMLmind XSL-FO Converter. Bioinformatics írta Antal, Péter, Hullám, Gábor, Millinghoffer, András, Hajós, Gergely, Marx, Péter, Arany, Ádám, Bolgár, Bence, Gézsi, András, Sárközy, Péter, és Poppe, László Publication date 2014 Szerzői jog © 2014 Antal Péter, Hullám Gábor, Millinghoffer András, Hajós Gergely, Marx Péter, Arany Ádám, Bolgár Bence, Gézsi András, Sárközy Péter, Poppe László Created by XMLmind XSL-FO Converter. Tartalom Bioinformatics .................................................................................................................................... 1 1. 1 DNA recombinant measurement technology, noise and error models ............................... 1 1.1. 1.1 Historic overview ............................................................................................... 1 1.1.1. 1.1.1 Clinical aspects of genome sequencing ............................................... 2 1.1.2. 1.1.2 Partial Genetic Association Studies .................................................... 2 1.1.3. 1.1.3 Genome Wide Association Studies ..................................................... 2 1.2. 1.2 First generation automated Sanger sequencing ................................................... 2 1.3. 1.3 Next generation sequencing technologies ........................................................... 3 1.3.1. 1.3.1 Pyrosequencing and pH based sequencing ......................................... -

Improved Approaches to Ligand Growing Through Fragment Docking and Fragment-Based Library Design
Improved approaches to ligand growing through fragment docking and fragment-based library design Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften (Dr. rer. nat.) dem Fachbereich Pharmazie der Philipps-Universit¨atMarburg vorgelegt von Florent Chevillard aus Toulouse Marburg/Lahn 2016 Erstgutachter Dr. Peter Kolb Institut f¨urPharmazeutische Chemie Philipps-Universit¨atMarburg Zweitgutachter Prof. Dr. Gerhard Klebe Institut f¨urPharmazeutische Chemie Philipps-Universit¨atMarburg Eingereicht am 28.7.2016 Tag der m¨undlichen Pr¨ufungam 23.9.2016 Hochschulkennziffer: 1180 i "And why do we fall, Bruce ? So we can learn to pick ourselves up." Thomas Wayne, Batman begins (2005) Abstract In the past two decades, fragment-based drug discovery (FBDD) has continuously gained popularity in drug discovery efforts and has become a dominant tool in order to explore novel chemical entities that might act as bioactive modulators. FBDD is intimately con- nected to fragment extension approaches, such as growing, merging or linking. These approaches can be accelerated using computational programs or semi-automated work- flows for de novo design. Although computers allow for the facile generation of millions of suggestions, this often comes at a price: uncertain synthetic feasibility of the generated compounds, potentially leading to a dead end in an optimization process. In this manuscript we developed two computational tools which could support the FBDD elaboration cycle: PINGUI and SCUBIDOO. PINGUI is a semi-automated workflow for fragments growing guided by both the protein structure and synthetic feasibility. SCUBIDOO is a freely accessible database which currently holds 21 M virtual products. This database was created by combining commercially available building blocks with robust organic reactions. -

Lobbying Contacts Report 1St January - 30Th June 2016
Conservative MEP Lobbying Contact Report 1st January - 30th June 2016 Member of European Richard ASHWORTH MEP Parliament Region South East England Date of Name of Client(s) Organisation/company Context contact lobbyist(s) - if applicable National Farmers Union of England 13/01/16 Fay Jones Agriculture and Wales (NFU) Polish food sector Małgorzata and Federation of 16/02/16 ContactEurope Agriculture Handzlik Agricultural Producers Union Richard Ali 16/02/16 FACE UK Hunting Tim Bonner 16/02/16 Gail Orton Tate and Lyle EU sugar market access National Farmers Union of England 17/02/16 Meurig Raymond Agriculture and Wales (NFU) National Farmers Union of England 12/04/16 Meurig Raymond Agriculture and Wales (NFU) Becky Thwaites 27/04/16 EU Dog & Cat Alliance Puppy smuggling and online sales of pets Anna Wade 03/05/16 Joe Stone Cargill Agricultural innovation National Farmers Union of England 03/05/16 Fay Jones Agriculture and Wales (NFU) Kelly Thomas ADS 01/06/16 Defence research Tim Lawrenson BAE Jim Miller Chip Councell Rosalind Leeck U.S. Soybean Export Council 15/06/16 Floyd Gaibler (USSEC) and the U.S. Grains EU-US Trade David Green Council (USGC) Benno van der Laan Name of European Parliamentary Intergroup, Positions of responsibility held club, forum or body of any kind with an Brief description of the nature of activity e.g. director, council member, or interest in European Parliamentary activities otherwise note as member A forum for discussion and information exchange The European Parliament Beer Club Member about issues that affect -

Conference Handbook 2013
genesis Conference Handbook 2013 2013 WELCOME TO EDINBURGH BIOQUARTER Designed to foster scientific creativity and encourage entrepreneurship among clinicians and researchers, BioQuarter combines world-class research from EDINBURGH LAUNCHES WORLD’S FIRST Edinburgh University’s College of Medicine and Veterinary Medicine with the long tradition of clinical VISUAL FIELD ANALYSER FOR CHILDREN excellence at NHS Lothian. Building one of Europe’s most In March 2012, Edinburgh significant concentrations of clinical and research assets, BioQuarter launched i2eye Diagnostics Limited, creators BioQuarter brings together a major teaching hospital, of the world’s first visual field medical school and three leading research institutes in analyser for children and hard- one location. Our seasoned business development team to-test patient groups. Based on research undertaken at the has spun out four new businesses and negotiated four University of Edinburgh and major industrial collaborations in less than two years, as NHS Lothian, this device enables well as generating 100 new product or business concepts young children’s field of vision to from its innovation competition. be tested accurately for the first the company has booked its first time, helping to pick up retinal orders and is set to take premises and optic nerve defects as well in our new incubator building, To find out more about how this commercialisation as certain forms of brain cancer. Nine the BioQuarter: partnership is building on Scotland’s successes Backed by private venture capital, www.i2eyediagnostics.com in life sciences, visit www.bioquarter.com Sponsors 3 Genesis 2013 Sponsors Contents Gold Sponsors Welcome 7 Keynote Speakers 8 - 9 Speaker Profiles 9 - 21 Programme 22-23 Floor Plan 24 Exhibitor List 25 Silver Sponsors The Genesis Talent Lounge 27 Exhibitor Profiles 31- 42 ON Hub 43 Corporate Patron Bronze Sponsors Corporate Sponsors Case Study Sponsors Partners Supporters Wifi Sponsor Media Partners Scotland. -

Press Release
Press release Embargoed until 10.00 (BST) Monday 11 May 2015 Leading medical experts recognised for excellence in research 48 researchers from across the UK have been recognised for their contribution to the advancement of medical science by election to the Fellowship of the Academy of Medical Sciences. Academy Fellows are elected for excellence in medical research, for innovative application of scientific knowledge or for their conspicuous service to healthcare. The expertise of the new Fellows includes addictions, anaesthesia, age-related diseases and animal biology. Professor Sir John Tooke PMedSci, President of the Academy of Medical Sciences said, “The Academy of Medical Sciences champions the excellence and diversity of medical science in the UK, and this is clearly demonstrated in this year’s cohort of new Fellows. Their broad range of expertise and fantastic achievements to date shows just how strong the Fellowship is – from the NHS knowledge of Sir Andrew Dillon to the policy experience of Professor Christl Donnelly. Their election is a much deserved honour, and I know they will contribute greatly to the Academy. I am delighted to welcome them all to the Fellowship, and look forward to working with them in the future.” This year, 17 (35%) of the new Fellows are women, compared to 23.2% of bioscience professors in the UK.* Professor Fiona Gribble FMedSci, elected this year, is a Wellcome Trust Senior Clinical Fellow at the University of Cambridge. Her research focuses on gastrointestinal endocrine cells, which are one of the main targets for developing new anti-diabetic and anti-obesity therapies. She has been presented with numerous awards to recognise her achievements, including the RD Lawrence lectureship, the Lister Prize, the Minkowski prize and the Viktor Mutt medal. -
Fragments, Hotspots and Target Identification: Decreasing Complexity in Drug Design
FRAGMENTS, HOTSPOTS AND TARGET IDENTIFICATION: DECREASING COMPLEXITY IN DRUG DESIGN Tom Blundell¸ Department of Biochemistry at Cambridge Abstract: Fragment-based drug discovery is a powerful and now widely used approach both in academia and industry to create novel high quality drug like molecules (Blundell et al., 2002; Murray & Rees (2009); Erlanson et al., 2016). This approach relies on screening a library consisting of small molecules (150-300 Da) against a target protein, using a variety of biochemical, biophysical and also structural biology methods. The low molecular weight of fragments represents a decrease of complexity and allows an efficient exploration of chemical space even when using small size libraries of around 1000 fragments. Although the fragments bind weakly, they tend to bind to hotspots forming well-defined interactions with the target protein. Thus, fragments can be subsequently elaborated into larger molecules with high affinity. An example of our approach to tuberculosis targets is found in Mendes and Blundell (2016) The approach developed in our academic laboratories in Cambridge builds on that developed in Astex, the company I cofounded in 1999 with Harren Jhoti and Chris Abell. It involves two stages. First we screen to identify hits using differential scanning fluorimetry, ligand-based one-dimensional 1H NMR spectroscopy and surface plasmon resonance (SPR). Second we determine the 3D-structures of protein-fragment complexes, followed by a study of thermodynamics using isothermal titration calorimetry (ITC) and kinetics of the binding process using again SPR Due to their small size fragments interact weakly with the target protein, usually between 0.1-5 mM, but those interactions tend to bind to hotspots that make large contributions to binding affinity. -
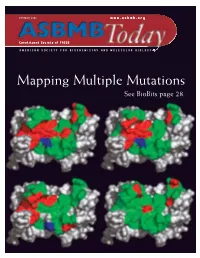
Mapping Multiple Mutations See Biobits Page 28
OCTOBER 2006 www.asbmb.org Constituent Society of FASEB AMERICAN SOCIETY FOR BIOCHEMISTRY AND MOLECULAR BIOLOGY Mapping Multiple Mutations See BioBits page 28. Why Choose 21 st Century Biochemicals For Custom Peptides and Antibodies? “…the dual phosphospecific antibody you “…of the 15 custom antibodies we ordered from your company, 13 worked. “…our in-house QC lab confirmed the Thanks for the great work!” high purity of the peptide.” Over 70 years combined experience manufacturing peptides and anti- bodies Put our expertise to work for you in a collaborative atmosphere The Pinnacle – Affinity Purified Antibodies >85% Success Rate! Peptide Sequencing Included for 100% Guaranteed Peptide Fidelity Complete 2 rabbit protocol with PhD technical support and $ 1675 epitope design ◆ From HPLC purified peptide to affinity purified antibody with no hidden charges!..........$1895 Special pricing through December 3 1, 2006 Phosphospecific Antibody Experts ◆ Custom Phosphospecific Ab’s as low as $2395 Come speak with our scientists at Neuroscience in Atlanta, GA - Booth 2127 and HUPO in Long Beach, CA - Booth 231 www.21stcenturybio.com 33 Locke Drive, Marlboro, MA 01752 Made in the P: 508.303.8222 Toll-free: 877.217.8238 U.S.A. F: 508.303.8333 E: [email protected] www.asbmb.org AMERICAN SOCIETY FOR BIOCHEMISTRY AND MOLECULAR BIOLOGY OCTOBER 2006 Volume 5, Issue 7 features 8 Osaka Bioscience Institute: A World Leader in Scientific Research 11 JLR Looks at Systems Biology 12 Education and Professional Development ON THE COVER: Human growth hormone 14 Judith Klinman to Receive ASBMB-Merck Award residues colored by contribution to receptor 15 Scott Emr Selected for ASBMB-Avanti Award recognition: red, favorable; 16 The Chromosome Cycle green, neutral; blue, unfavorable.