<I>Corynebacterium Bovis</I>
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Corynebacterium Species Rarely Cause Orthopedic Infections
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2018 Corynebacterium species rarely cause orthopedic infections Kalt, Fabian ; Schulthess, Bettina ; Sidler, Fabian ; Herren, Sebastian ; Fucentese, Sandro F ; Zingg, Patrick O ; Berli, Martin ; Zinkernagel, Annelies S ; Zbinden, Reinhard ; Achermann, Yvonne Abstract: Corynebacterium spp. are rarely considered as pathogens but data in orthopedic infections are sparse. Therefore, we asked how often Corynebacterium spp. caused an infection in a defined cohort of orthopedic patients with a positive culture. In addition, we aimed to determine the species variety and susceptibility of isolated strains in regards to potential treatment strategies. Between 2006 and 2015, we retrospectively assessed all Corynebacterium sp. bone and joint cultures from an orthopedic ward. The isolates were considered as relevant indicating an infection if the same Corynebacterium sp. was present in at least two samples. We found 97 orthopedic cases with isolation of Corynebacterium spp. (128 positive samples), mainly Corynebacterium tuberculostearicum (n=26), Corynebacterium amycolatum (n=17), Corynebacterium striatum (n=13), and Corynebacterium afermentans (n=11). Compared to a cohort of positive blood cultures, we found significantly more C. striatum and C. tuberculostearicum but no C. jeikeium cases. Only 16 cases out 66 cases (24.2%) with an available diagnostic set of at least 2 samples had an infection. Antibiotic susceptibility testing (AST) of different antibiotics showed various susceptibility results except for vancomycin and linezolid with a 100% susceptibility rate. Rates of susceptibility of corynebacteria isolated from orthopedic samples and of isolates from blood cultures were comparable. In conclusion, our study results confirmed that Corynebacterium sp. -

Corynebacterium Bovis: a Rare Case of Persistent Bacterial Keratitis and Corneal Perforation
Open Access Case Report DOI: 10.7759/cureus.16913 Corynebacterium Bovis: A Rare Case of Persistent Bacterial Keratitis and Corneal Perforation Mohammed Elsheikh 1 , Ahmed Elsayed 1 , Nicholas Bennett 1 , Martin Connor 2 1. Ophthalmology, NHS Dumfries & Galloway, Dumfries, GBR 2. Microbiology, NHS Dumfries & Galloway, Dumfries, GBR Corresponding author: Mohammed Elsheikh, [email protected] Abstract We report a rare case of severe, non-contact lens-related Corynebacterium bovis corneal infection on a background of viral keratitis, resulting in corneal abscess formation with subsequent corneal perforation. An 89-year-old Caucasian lady presented with a significant epithelial defect and a dense stromal infiltrates on a background of herpes zoster keratitis, ultimately resulting in corneal perforation. Enrichment culture obtained from corneal scraping isolated the unusual organism Corynebacterium bovis. This was treated with a combination of culture-directed, targeted course of antibiotics and surgical interventions. To the best of our knowledge, this is the first reported case of profuse bacterial keratitis secondary to Corynebacterium bovis infiltration, on a background of viral keratitis, resulting in corneal abscess formation and subsequent perforation. This report highlights this rare bacterium’s characteristics including its pathogenicity in causing severe corneal disease, particularly in immunosuppressed environments such as in this case, apparent antibiotic sensitivities & resistance, and potential transmission route. Categories: Ophthalmology, Infectious Disease Keywords: corynebacterium bovis, cornea, microbial keratitis, corneal perforation, corneal infection Introduction Corynebacterium species are frequently isolated on the conjunctiva of healthy adults and are thus increasingly regarded as non-pathogenic bacteria [1,2]. Recently, however, there have been increasing reports detailing the pathogenicity of Corynebacterium species isolated on ocular surfaces, with immunosuppression representing a risk factor [3-6]. -

( 12 ) United States Patent
US009956282B2 (12 ) United States Patent ( 10 ) Patent No. : US 9 ,956 , 282 B2 Cook et al. (45 ) Date of Patent: May 1 , 2018 ( 54 ) BACTERIAL COMPOSITIONS AND (58 ) Field of Classification Search METHODS OF USE THEREOF FOR None TREATMENT OF IMMUNE SYSTEM See application file for complete search history . DISORDERS ( 56 ) References Cited (71 ) Applicant : Seres Therapeutics , Inc. , Cambridge , U . S . PATENT DOCUMENTS MA (US ) 3 ,009 , 864 A 11 / 1961 Gordon - Aldterton et al . 3 , 228 , 838 A 1 / 1966 Rinfret (72 ) Inventors : David N . Cook , Brooklyn , NY (US ) ; 3 ,608 ,030 A 11/ 1971 Grant David Arthur Berry , Brookline, MA 4 ,077 , 227 A 3 / 1978 Larson 4 ,205 , 132 A 5 / 1980 Sandine (US ) ; Geoffrey von Maltzahn , Boston , 4 ,655 , 047 A 4 / 1987 Temple MA (US ) ; Matthew R . Henn , 4 ,689 ,226 A 8 / 1987 Nurmi Somerville , MA (US ) ; Han Zhang , 4 ,839 , 281 A 6 / 1989 Gorbach et al. Oakton , VA (US ); Brian Goodman , 5 , 196 , 205 A 3 / 1993 Borody 5 , 425 , 951 A 6 / 1995 Goodrich Boston , MA (US ) 5 ,436 , 002 A 7 / 1995 Payne 5 ,443 , 826 A 8 / 1995 Borody ( 73 ) Assignee : Seres Therapeutics , Inc. , Cambridge , 5 ,599 ,795 A 2 / 1997 McCann 5 . 648 , 206 A 7 / 1997 Goodrich MA (US ) 5 , 951 , 977 A 9 / 1999 Nisbet et al. 5 , 965 , 128 A 10 / 1999 Doyle et al. ( * ) Notice : Subject to any disclaimer , the term of this 6 ,589 , 771 B1 7 /2003 Marshall patent is extended or adjusted under 35 6 , 645 , 530 B1 . 11 /2003 Borody U . -

Septicaemiain a Granulocytopenic Patientcaused By
- Postgraduate Medical Journal (1989) 65, 247 248 Postgrad Med J: first published as 10.1136/pgmj.65.762.247 on 1 April 1989. Downloaded from Septicaemia in a granulocytopenic patient caused by Corynebacterium striatum Lawrence Dall', William G. Barnes2 and Dennis Hurford' 'Department ofMedicine, University ofMissouri, School ofMedicine-Kansas City, 2411 Holmes Street, Kansas City, Missouri 64108 and2Department ofPathology, Truman Medical Center, and School ofMedicine, University ofMissouri-Kansas City, 2301 Holmes Street, Kansas City, Missouri 64108, USA. Summary: A 64 year old woman with metastatic endometrial carcinoma was admitted to the hospital after three grand mal seizures. Blood cultures yielded Corynebacterium striatum. The patient responded to parenteral ampicillin therapy. This is believed to be the first case of sepsis caused by this organism. Introduction Neutropenic patients are susceptible to a wide variety platelet count 95 x 109/1, and white blood cell count of infections from unusual organisms. The incidence 0.99 x 109/l (48% polymorphonuclear leucocytes). of bacteraemia increases as the neutrophil count falls Chest X-ray, and cranial computed tomography below 500 x 106/1 with most of the organisms revealed multiple metastatic lesions. Two sets ofblood originating from the patient's endogenous flora. The cultures (4/4 bottles) obtained on admission revealed a organism implicated in this case, Corynebacterium nonsporulating, aerobic, weakly haemolytic, non- striatum, is recognized as a part of the microbial flora motile, catalase positive, pleomorphic diptheroid-like, of the human skin and nasal mucous membranes. Gram-positive bacillus which fermented glucose, fruc- We believe this report to be the first documented tose, mannose, lactose and maltose. -

Detection of Corynebacterium Bovis Infection in Athymic Nude Mice from a Research Animal Facility in Korea
pISSN 1229-845X, eISSN 1976-555X JOURNAL OF J. Vet. Sci. (2014), 15(4), 583-586 http://dx.doi.org/10.4142/jvs.2014.15.4.583 Veterinary Received: 5 Feb. 2014, Revised: 8 May 2014, Accepted: 19 Jun. 2014 Science Case Report Detection of Corynebacterium bovis infection in athymic nude mice from a research animal facility in Korea Tae-Hyoun Kim1,2, Dong-Su Kim1, Ju-Hee Han1,2, Seo-Na Chang1,2, Kyung-Sul Kim1, Seung-Hyeok Seok3, Dong-Jae Kim4, Jong-Hwan Park4, Jae-Hak Park1,2,* 1Department of Laboratory Animal Medicine, and 2BK21 PLUS Program for Creative Veterinary Science Research, Research Institute for Veterinary, College of Veterinary Medicine, Seoul National University, Seoul 151-742, Korea 3Department of Microbiology and Immunology, Seoul National University College of Medicine, Seoul 110-799, Korea 4Department of Biochemistry, College of Medicine, Konyang University, Daejeon 302-832, Korea Corynebacterium (C.) bovis infection in nude mice causes examination, Test mouse 1 did not present any clinical hyperkeratosis and weight loss and has been reported signs. In contrast, Test mouse 2 and Test mouse 3 had worldwide but not in Korea. In 2011, nude mice from an yellow-white scales on the dorsal skin (Fig. 1), and their animal facility in Korea were found to have white flakes on weight was lower than that suggested by the vendor (Table their dorsal skin. Histopathological testing revealed that the 1). For further diagnosis, matched healthy control mice mice had hyperkeratosis and Gram-positive bacteria were (Central Lab Animal, Korea) were purchased and included found in the skin. -

Antibiotic Susceptibility of Major Bacteria Cause Ovine Mastitis in River Nile State, Sudan
Imperial Journal of Interdisciplinary Research (IJIR) Vol-3, Issue-1, 2017 ISSN: 2454-1362, http://www.onlinejournal.in Antibiotic Susceptibility of Major Bacteria Cause Ovine Mastitis in River Nile State, Sudan Fadlalla E.2 ,1٭Abubaker A. El Ayis 1Department of Medicine, College of Veterinary Medicine, Bahri University (Sudan). 2Department of Biochemistry, College of Veterinary Medicine, Bahri University (Sudan). Abstract: In this study a total of 40 milk samples Procaine Penicillin and Cloxacillin. 50.0% of positive for California Mastitis Test (CMT) were Klebsiella pneumoniae isolates were sensitive to collected from ewes in the three localities of River Ciprofloxacin, Gentamycin and Kanamycin. All Nile State (Atbara, Barbar and El Damer). Samples strains were resistant to Ampicillin, Cephalothin, were submitted for bacteriological examination. Chloramphenicol, Erythromycin Procaine The aerobic bacteria isolated and identified from Penicillin, Cloxacillin and Tetracycline. All ewes’ milk samples were 19 Staphylococci Enterobacter spp. Isolates were sensetive to (21.0%), 9 Streptococci (9.9%), 5 Echerichia coli Cephalothin and Chloramphenicol, 66.7% to (5.6%), 4 Corynebacterium bovis (3.3%), 3 Ciprofloxacin, Tetracycline and Gentamycin and Enterobacter spp. (3.3%), 2 Klebsiella pneumoniae all isolates were resstant to Ampicillin, spp, (2.2%) and 2 Enterococcus faecalis (2.2%). Erythromycin, Kanamycin, Procaine Penicillin and All isolates were subjected to sensitivity tests using Cloxacillin. 10 antibiotics in use for treatment of mastitis in Sudan. 89.5% Staphylococcal isolates were I. INTRODUCTION sensitive to Ampicillin, 73.3% to Cephalothin, 68.4% to Ciprofloxacin and Kanamycin, 63.2% to Mastitis is one of the more common health Gentamycin, 57.9% to Chloramphenicol, 26.3% to problems affecting sheep and goats. -
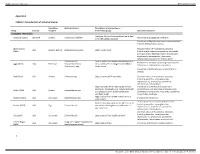
Complete Restriction
Supplementary material BMJ Global Health Appendix 1 Table S1: Characteristics of individual studies Population Bacteria studied Description of intervention or Study Country sampled intervention group Outcomes measured Complete restriction Ecological farms with no antibiotic use in feed Aarestrup (1995) Denmark Broilers Enterococcus faecium Resistance to glycopeptide antibiotics or for therapeutic purposes Prevalence of Staphylococcus aureus and methicillin resistant Staphylococcus aureus Abdalrahman Characterization of resistance to antibiotics USA Broilers, turkeys Staphylococcus aureus Organic retail meat (2015) (including beta-lactams, tetracyclines, macrolides, aminoglycosides, fluoroquinolones, lincosamides, amphenicols, glycopeptides, rifamycines, sulfonamides) and presence of toxin genes Escherichia coli Pasture-raised non-antimicrobial treated beef Resistance to antibiotics (including cephalosporins, Agga (2015) USA Beef cows Salmonella enterica cows, which were co-mingled with ceftiofur tetracyclines, sulfonamides, macrolides) Enterococcus spp. treated cows Prevalence of Salmonella spp. in fecal and farm samples Alali (2010) USA Broilers Salmonella spp. Organic farms (USDA certified) Characterization of resistance to antibiotics (including penicillins, aminoglycosides, cephalosporins, amphenicols, quinolones, sulfonamides, tetracyclines) Organic broilers which were raised without Prevalence and antimicrobial resistance of Campylobacter spp. (including aminoglycosides, Bailey (2019) USA Broilers Campylobacter spp. antibiotics, -

Udder Pathogens and Their Resistance to Antimicrobial Agents in Dairy Cows in Estonia Piret Kalmus1*†, Birgit Aasmäe2†, Age Kärssin3†, Toomas Orro2†, Kalle Kask1†
Kalmus et al. Acta Veterinaria Scandinavica 2011, 53:4 http://www.actavetscand.com/content/53/1/4 RESEARCH Open Access Udder pathogens and their resistance to antimicrobial agents in dairy cows in Estonia Piret Kalmus1*†, Birgit Aasmäe2†, Age Kärssin3†, Toomas Orro2†, Kalle Kask1† Abstract Background: The goal of this study was to estimate the distribution of udder pathogens and their antibiotic resistance in Estonia during the years 2007-2009. Methods: The bacteriological findings reported in this study originate from quarter milk samples collected from cows on Estonian dairy farms that had clinical or subclinical mastitis. The samples were submitted by local veterinarians to the Estonian Veterinary and Food Laboratory during 2007-2009. Milk samples were examined by conventional bacteriology. In vitro antimicrobial susceptibility testing was performed with the disc diffusion test. Logistic regression with a random herd effect to control for clustering was used for statistical analysis. Results: During the study period, 3058 clinical mastitis samples from 190 farms and 5146 subclinical mastitis samples from 274 farms were investigated. Positive results were found in 57% of the samples (4680 out of 8204), and the proportion did not differ according to year (p > 0.05). The proportion of bacteriologically negative samples was 22.3% and that of mixed growth was 20.6%. Streptococcus uberis (Str. uberis) was the bacterium isolated most frequently (18.4%) from cases of clinical mastitis, followed by Escherichia coli (E. coli) (15.9%) and Streptococcus agalactiae (Str. agalactiae) (11.9%). The bacteria that caused subclinical mastitis were mainly Staphylococcus aureus (S. aureus) (20%) and coagulase-negative staphylococci (CNS) (15.4%). -

The Effect of Media Composition on Nitrile Hydratase Activity and Stability, and on Cell Envelope Components of Rhodococcus DAP 96253
Georgia State University ScholarWorks @ Georgia State University Biology Dissertations Department of Biology 11-30-2008 The Effect of Media Composition on Nitrile Hydratase Activity and Stability, and on Cell Envelope Components of Rhodococcus DAP 96253 Trudy-Ann Marie Tucker Follow this and additional works at: https://scholarworks.gsu.edu/biology_diss Part of the Biology Commons Recommended Citation Tucker, Trudy-Ann Marie, "The Effect of Media Composition on Nitrile Hydratase Activity and Stability, and on Cell Envelope Components of Rhodococcus DAP 96253." Dissertation, Georgia State University, 2008. https://scholarworks.gsu.edu/biology_diss/56 This Dissertation is brought to you for free and open access by the Department of Biology at ScholarWorks @ Georgia State University. It has been accepted for inclusion in Biology Dissertations by an authorized administrator of ScholarWorks @ Georgia State University. For more information, please contact [email protected]. EFFECT OF CHANGES IN MEDIA COMPOSITION ON NITRILE HYDRATASE ACTIVITY AND STABILITY AND ON CELL ENVELOPE COMPONENTS OF RHODOCOCCUS SP DAP 96253 by TRUDY-ANN TUCKER Under the Direction of George E. Pierce ABSTRACT Rhodococcus is an important industrial organism that possesses diverse metabolic capabilities, it also has a unique cell envelope, composed of an outer layer of mycolic acids and glycolipids (free or bound lipids generally linked to the sugar trehalose). Rhodococcus is able to transform nitriles to the corresponding amide by the enzyme Nitrile Hydratase (NHase), therefore rhodococcal cells can be utilized as biocatalysts in the detoxification of nitrile waste water or in the production of industrially important amides such as acrylamide. However, the NHase within the native cells must be stable with high activity. -

An Overview on the Epidemiology and Diagnosis of Bovine Mastitis
Advances in Life Science and Technology www.iiste.org ISSN 2224-7181 (Paper) ISSN 2225-062X (Online) Vol.35, 2015 An Overview on the Epidemiology and Diagnosis of Bovine Mastitis Eyob Eshetu School of Veterinary Medicine, Wolaita Sodo University, Ethiopia Introduction Mastitis is the most common infectious disease encountered in intensively –farmed dairy cattle (Quinn et al ., 2002). It is inflammation of the parenchyma of the mammary gland regardless of the cause. Mastitis is therefore characterized by a range of physical and chemical change in the milk include discoloration, the present of clot and present of large number of leukocyte (Radostits et al ., 2007). In other words, bovine mastitis a response to ascending infection of the gland by the way of the teat canal (Jubb et al ., 1993). There is swelling, heat, pain and edema in the mammary gland in many clinical cases. However, a large proportion of mastitic gland is not readily detectable by manual palpation or by visual examination of the milk strip cup; these quarters represent subclinical infection (Radostits et al ., 2007). Etiology A total of about 140 microbial species, sub species and serovars have been isolated from the bovine mammary gland. However a relatively small number of them are responsible for most cases of mastitis (Quinn et al ., 2002 and Radostits et al., 2007). The bovine mammary gland is the principal reservoir of infectious agent which causes contagious mastitis, namely Staphylococcus aureus , Streptococcus agalactiae, Mycoplasma bovis and corynebacterium bovis (Quinn et al ., 2002). Staphylococcus aureus is one of the most causes of bovine subclinical mastitis (Wilson et al ., 1997). -

List of Contents
Biochemical and biophysical characterization of various cell wall channel-forming proteins By Nafiseh Soltanmohammadi Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biochemical Engineering Approved, thesis committee Prof. Dr. Roland Benz Prof. Dr. Mathias Winterhalter Prof. Dr. Miquel Viñas Ciordia Date of defense: March 22, 2013 School of engineering and science Abstract The mycolic-acid layer of certain gram-positive bacteria, the mycolata, represents an additional permeability barrier for the permeation of small water-soluble solutes. Consequently, it was shown in recent years that the mycolic acid layer of individual bacteria of the group mycolata contains pores, called porins, for the passage of hydrophilic solutes. Corynebacterium amycolatum, a pathogenic Corynebacterium species, belongs to the Corynebacteriaceae family but it lacks corynomycolic acids in its cell wall. Despite the absence of corynomycolic acids the cell wall of C. amycolatum contains a cation-selective cell wall channel, which may be responsible for the limited permeability of the cell wall of C. amycolatum. In the first project of this thesis and based on partial sequencing of the protein responsible for channel formation derived from C. amycolatum ATCC 49368 we were able to identify the corresponding gene coram0001_1986 within the known genome sequence of C. amycolatum SK46 that codes for the cell wall channel. The corresponding gene of C. amycolatum ATCC 49368 was cloned into the plasmid pXHis for its expression in Corynebacterium glutamicum ∆porA∆porH. Biophysical characterization of the purified protein (PorAcoram) suggested that coram0001_1986 is indeed the gene coding for the pore-forming protein PorAcoram in C. -

Corynebacterium Bovis Shoulder Prosthetic Joint Infection
Available online at www.sciencedirect.com Diagnostic Microbiology and Infectious Disease 64 (2009) 213–215 www.elsevier.com/locate/diagmicrobio Case Reports Corynebacterium bovis shoulder prosthetic joint infection: the first reported case ⁎ Yvonne Achermanna, , Andrej Trampuzb, Fabricio Moroc, Jürg Wüstd, Markus Vogta aDepartment of Medicine, Cantonal Hospital Zug, CH-6340 Baar, Switzerland bInfectious Diseases Service, Department of Medicine, Centre Hospitalier Universitaire Vaudois (CHUV) and University of Lausanne, CH-1011 Lausanne, Switzerland cUpper Extremity Department, Schulthessclinic, CH-8008 Zurich, Switzerland dUNILABS Zürich, CH-8034 Zurich, Switzerland Received 11 January 2009; accepted 4 February 2009 Abstract We report the first case of Corynebacterium bovis shoulder prosthetic joint infection. The organism was isolated from intraoperative tissue culture and from the removed prosthesis using sonication. A 2-stage exchange and 3 months of antibiotic therapy were performed. C. bovis may cause implant-associated infections, which can manifest as low-grade infection. © 2009 Elsevier Inc. All rights reserved. Keywords: Prosthetic joint infection; Corynebacterium bovis; Shoulder prosthesis 1. Introduction in whom C. bovis with identical phenotypic characteristics was repeatedly isolated from different specimens collected 2 Corynebacterium bovis is a rare human pathogen (Coyle weeks apart. and Lipsky, 1990). It is a common commensal of the bovine udder and may cause bovine mastitis. Although the mode of 2. Case report transmission to humans is unknown, drinking contaminated bovine milk and nosocomial acquisition is discussed (Smith A 62-year-old female developed secondary arthrosis of and Locksley, 1982). Until know, 8 patients with C. bovis the acromioclavicular joint after surgical stabilization of the infections were described, including line-related sepsis, left shoulder 9 years before presentation.