Tracking the Embryonic Stem Cell Transition from Ground State Pluripotency
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-
Stem Cell Factor Is Selectively Secreted by Arterial Endothelial Cells in Bone Marrow
ARTICLE DOI: 10.1038/s41467-018-04726-3 OPEN Stem cell factor is selectively secreted by arterial endothelial cells in bone marrow Chunliang Xu1,2, Xin Gao1,2, Qiaozhi Wei1,2, Fumio Nakahara 1,2, Samuel E. Zimmerman3,4, Jessica Mar3,4 & Paul S. Frenette 1,2,5 Endothelial cells (ECs) contribute to haematopoietic stem cell (HSC) maintenance in bone marrow, but the differential contributions of EC subtypes remain unknown, owing to the lack 1234567890():,; of methods to separate with high purity arterial endothelial cells (AECs) from sinusoidal endothelial cells (SECs). Here we show that the combination of podoplanin (PDPN) and Sca-1 expression distinguishes AECs (CD45− Ter119− Sca-1bright PDPN−) from SECs (CD45− Ter119− Sca-1dim PDPN+). PDPN can be substituted for antibodies against the adhesion molecules ICAM1 or E-selectin. Unexpectedly, prospective isolation reveals that AECs secrete nearly all detectable EC-derived stem cell factors (SCF). Genetic deletion of Scf in AECs, but not SECs, significantly reduced functional HSCs. Lineage-tracing analyses suggest that AECs and SECs self-regenerate independently after severe genotoxic insults, indicating the per- sistence of, and recovery from, radio-resistant pre-specified EC precursors. AEC-derived SCF also promotes HSC recovery after myeloablation. These results thus uncover heterogeneity in the contribution of ECs in stem cell niches. 1 The Ruth L. and David S. Gottesman Institute for Stem Cell and Regenerative Medicine Research, Albert Einstein College of Medicine, New York, NY 10461, USA. 2 Department of Cell Biology, Albert Einstein College of Medicine, New York, NY 10461, USA. 3 Department of Systems and Computational Biology, Albert Einstein College of Medicine, New York, NY 10461, USA. -

Goat Anti-JAM2 / JAMB / CD322 Antibody Peptide-Affinity Purified Goat Antibody Catalog # Af1576a
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Goat Anti-JAM2 / JAMB / CD322 Antibody Peptide-affinity purified goat antibody Catalog # AF1576a Specification Goat Anti-JAM2 / JAMB / CD322 Antibody - Product Information Application WB Primary Accession P57087 Other Accession NP_067042, 58494, 67374 (mouse), 619374 (rat) Reactivity Human Predicted Mouse, Rat, Dog, Cow Host Goat Clonality Polyclonal Concentration 100ug/200ul Isotype IgG Calculated MW 33207 Goat Anti-JAM2 / JAMB / CD322 Antibody - AF1576a (0.5 µg/ml) staining of Human Additional Information Heart lysate (35 µg protein in RIPA buffer) with (B) and without (A) blocking with the Gene ID 58494 immunising peptide. Primary incubation was 1 hour. Detected by chemiluminescence. Other Names Junctional adhesion molecule B, JAM-B, Junctional adhesion molecule 2, JAM-2, Goat Anti-JAM2 / JAMB / CD322 Antibody - Vascular endothelial junction-associated Background molecule, VE-JAM, CD322, JAM2, C21orf43, VEJAM Tight junctions represent one mode of cell-to-cell adhesion in epithelial or endothelial Format cell sheets, forming continuous seals around 0.5 mg IgG/ml in Tris saline (20mM Tris cells and serving as a physical barrier to pH7.3, 150mM NaCl), 0.02% sodium azide, prevent solutes and water from passing freely with 0.5% bovine serum albumin through the paracellular space. The protein encoded by this immunoglobulin superfamily Storage gene member is localized in the tight junctions Maintain refrigerated at 2-8°C for up to 6 between high endothelial cells. It acts as an months. For long term storage store at adhesive ligand for interacting with a variety of -20°C in small aliquots to prevent immune cell types and may play a role in freeze-thaw cycles. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

CD Markers Are Routinely Used for the Immunophenotyping of Cells
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called CD markers are routinely used for the immunophenotyping of cells. Despite this use, they are not limited to roles in the immune system and perform a variety of roles in cell differentiation, adhesion, migration, blood clotting, gamete fertilization, amino acid transport and apoptosis, among many others. As such, Proteintech’s mini catalog featuring its antibodies targeting CD markers is applicable to a wide range of research disciplines. PRODUCT FOCUS PECAM1 Platelet endothelial cell adhesion of blood vessels – making up a large portion molecule-1 (PECAM1), also known as cluster of its intracellular junctions. PECAM-1 is also CD Number of differentiation 31 (CD31), is a member of present on the surface of hematopoietic the immunoglobulin gene superfamily of cell cells and immune cells including platelets, CD31 adhesion molecules. It is highly expressed monocytes, neutrophils, natural killer cells, on the surface of the endothelium – the thin megakaryocytes and some types of T-cell. Catalog Number layer of endothelial cells lining the interior 11256-1-AP Type Rabbit Polyclonal Applications ELISA, FC, IF, IHC, IP, WB 16 Publications Immunohistochemical of paraffin-embedded Figure 1: Immunofluorescence staining human hepatocirrhosis using PECAM1, CD31 of PECAM1 (11256-1-AP), Alexa 488 goat antibody (11265-1-AP) at a dilution of 1:50 anti-rabbit (green), and smooth muscle KD/KO Validated (40x objective). alpha-actin (red), courtesy of Nicola Smart. PECAM1: Customer Testimonial Nicola Smart, a cardiovascular researcher “As you can see [the immunostaining] is and a group leader at the University of extremely clean and specific [and] displays Oxford, has said of the PECAM1 antibody strong intercellular junction expression, (11265-1-AP) that it “worked beautifully as expected for a cell adhesion molecule.” on every occasion I’ve tried it.” Proteintech thanks Dr. -

Signals of the Neuropilin-1–MET Axis and Cues of Mechanical Force Exertion Converge to Elicit Inflammatory Activation in Coherent Endothelial Cells
Signals of the Neuropilin-1−MET Axis and Cues of Mechanical Force Exertion Converge to Elicit Inflammatory Activation in Coherent Endothelial Cells This information is current as of September 27, 2021. Maryam Rezaei, Ana C. Martins Cavaco, Jochen Seebach, Stephan Niland, Jana Zimmermann, Eva-Maria Hanschmann, Rupert Hallmann, Hermann Schillers and Johannes A. Eble J Immunol published online 28 January 2019 Downloaded from http://www.jimmunol.org/content/early/2019/01/27/jimmun ol.1801346 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2019/01/27/jimmunol.180134 Material 6.DCSupplemental Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 27, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2019 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published January 28, 2019, doi:10.4049/jimmunol.1801346 The Journal of Immunology Signals of the Neuropilin-1–MET Axis and Cues of Mechanical Force Exertion Converge to Elicit Inflammatory Activation in Coherent Endothelial Cells Maryam Rezaei,* Ana C. -

Network-Based Genetic Profiling, and Therapeutic Target Identification Of
bioRxiv preprint doi: https://doi.org/10.1101/480632; this version posted November 29, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Network-based genetic profiling, and therapeutic target identification of Thyroid Cancer 1st Md. Ali Hossain 2nd Tania Akter Asa 3rd Julian Quinn 4rdMd. Mijanur Rahman Dept. of CSE Dept. of EEE Bone biology divisions Dept. of CSE MIU, Jahangirnagar University Islamic University Garvan Institute of Medical Research JKKNIU Dhaka, Bangladesh Kushtia, Bangladesh Sydney, Australia Bangladesh ali:cse:bd@gmail:com tania:eee:iu@gmail:com j:quinn@garvan:org:au mijan cse@yahoo:com 5th Fazlul Huq 6th Mohammad Ali Moni Biomedical Sciences, Faculty of Medicine and Health Biomedical Sciences, Faculty of Medicine and Health The University of Sydney The University of Sydney Sydney, Australia Sydney, Australia fazlul:huq@sydney:edu:au mohammad:moni@sydney:edu:au Abstract—Molecular mechanisms that underlie the pathogene- I. INTRODUCTION sis and progression of malignant thyroid cancer (TC) are poorly understood. In this study, we employed network-based integrative Thyroid cancer (TC) is the most common endocrine ma- analyses of TC lesions to identify key molecules that are possible lignant disease [24], and although it has a relatively low hub genes and proteins in molecular pathways active in TC. We thus studied a microarray gene expression dataset (GSE82208, mortality rate compared to most other common metastatic n=52) that compared TC to benign thyroid tumours (follicular diseases its incidence is rising and is now the fifth most thyroid adenomas) in order to identify differentially expressed common cancer disease in women, notably affecting those genes (DEGs) in TC. -

Supplementary Table 1: List of the 316 Genes Regulated During Hyperglycemic Euinsulinemic Clamp in Skeletal Muscle
Supplementary Table 1: List of the 316 genes regulated during hyperglycemic euinsulinemic clamp in skeletal muscle. UGCluster Name Symbol Fold Change Cytoband Response to stress Hs.517581 Heme oxygenase (decycling) 1 HMOX1 3.80 22q12 Hs.374950 Metallothionein 1X MT1X 2.20 16q13 Hs.460867 Metallothionein 1B (functional) MT1B 1.70 16q13 Hs.148778 Oxidation resistance 1 OXR1 1.60 8q23 Hs.513626 Metallothionein 1F (functional) MT1F 1.47 16q13 Hs.534330 Metallothionein 2A MT2A 1.45 16q13 Hs.438462 Metallothionein 1H MT1H 1.42 16q13 Hs.523836 Glutathione S-transferase pi GSTP1 -1.74 11q13 Hs.459952 Stannin SNN -1.92 16p13 Immune response, cytokines & related Hs.478275 TNF (ligand) superfamily, member 10 (TRAIL) TNFSF10 1.58 3q26 Hs.278573 CD59 antigen p18-20 (protectin) CD59 1.49 11p13 Hs.534847 Complement component 4B, telomeric C4A 1.47 6p21.3 Hs.535668 Immunoglobulin lambda variable 6-57 IGLV6-57 1.40 22q11.2 Hs.529846 Calcium modulating ligand CAMLG -1.40 5q23 Hs.193516 B-cell CLL/lymphoma 10 BCL10 -1.40 1p22 Hs.840 Indoleamine-pyrrole 2,3 dioxygenase INDO -1.40 8p12-p11 Hs.201083 Mal, T-cell differentiation protein 2 MAL2 -1.44 Hs.522805 CD99 antigen-like 2 CD99L2 -1.45 Xq28 Hs.50002 Chemokine (C-C motif) ligand 19 CCL19 -1.45 9p13 Hs.350268 Interferon regulatory factor 2 binding protein 2 IRF2BP2 -1.47 1q42.3 Hs.567249 Contactin 1 CNTN1 -1.47 12q11-q12 Hs.132807 MHC class I mRNA fragment 3.8-1 3.8-1 -1.48 6p21.3 Hs.416925 Carcinoembryonic antigen-related cell adhesion molecule 19 CEACAM19 -1.49 19q13.31 Hs.89546 Selectin E (endothelial -

Differences in Gene Expression Profiles and Carcinogenesis Pathways Involved in Cisplatin Resistance of Four Types of Cancer
596 ONCOLOGY REPORTS 30: 596-614, 2013 Differences in gene expression profiles and carcinogenesis pathways involved in cisplatin resistance of four types of cancer YONG YANG1,2, HUI LI1,2, SHENGCAI HOU1,2, BIN HU1,2, JIE LIU1,3 and JUN WANG1,3 1Beijing Key Laboratory of Respiratory and Pulmonary Circulation, Capital Medical University, Beijing 100069; 2Department of Thoracic Surgery, Beijing Chao-Yang Hospital, Capital Medical University, Beijing 100020; 3Department of Physiology, Capital Medical University, Beijing 100069, P.R. China Received December 23, 2012; Accepted March 4, 2013 DOI: 10.3892/or.2013.2514 Abstract. Cisplatin-based chemotherapy is the standard Introduction therapy used for the treatment of several types of cancer. However, its efficacy is largely limited by the acquired drug Cisplatin is primarily effective through DNA damage and is resistance. To date, little is known about the RNA expression widely used for the treatment of several types of cancer, such changes in cisplatin-resistant cancers. Identification of the as testicular, lung and ovarian cancer. However, the ability RNAs related to cisplatin resistance may provide specific of cancer cells to become resistant to cisplatin remains a insight into cancer therapy. In the present study, expression significant impediment to successful chemotherapy. Although profiling of 7 cancer cell lines was performed using oligo- previous studies have identified numerous mechanisms in nucleotide microarray analysis data obtained from the GEO cisplatin resistance, it remains a major problem that severely database. Bioinformatic analyses such as the Gene Ontology limits the usefulness of this chemotherapeutic agent. Therefore, (GO) and KEGG pathway were used to identify genes and it is crucial to examine more elaborate mechanisms of cisplatin pathways specifically associated with cisplatin resistance. -

Mouse CD Marker Chart Bdbiosciences.Com/Cdmarkers
BD Mouse CD Marker Chart bdbiosciences.com/cdmarkers 23-12400-01 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD1d CD1.1, CD1.2, Ly-38 Lipid, Glycolipid Ag + + + + + + + + CD104 Integrin b4 Laminin, Plectin + DNAX accessory molecule 1 (DNAM-1), Platelet and T cell CD226 activation antigen 1 (PTA-1), T lineage-specific activation antigen 1 CD112, CD155, LFA-1 + + + + + – + – – CD2 LFA-2, Ly-37, Ly37 CD48, CD58, CD59, CD15 + + + + + CD105 Endoglin TGF-b + + antigen (TLiSA1) Mucin 1 (MUC1, MUC-1), DF3 antigen, H23 antigen, PUM, PEM, CD227 CD54, CD169, Selectins; Grb2, β-Catenin, GSK-3β CD3g CD3g, CD3 g chain, T3g TCR complex + CD106 VCAM-1 VLA-4 + + EMA, Tumor-associated mucin, Episialin + + + + + + Melanotransferrin (MT, MTF1), p97 Melanoma antigen CD3d CD3d, CD3 d chain, T3d TCR complex + CD107a LAMP-1 Collagen, Laminin, Fibronectin + + + CD228 Iron, Plasminogen, pro-UPA (p97, MAP97), Mfi2, gp95 + + CD3e CD3e, CD3 e chain, CD3, T3e TCR complex + + CD107b LAMP-2, LGP-96, LAMP-B + + Lymphocyte antigen 9 (Ly9), -
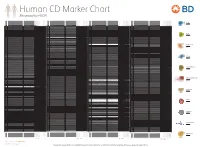
Reviewed by HLDA1
Human CD Marker Chart Reviewed by HLDA1 T Cell Key Markers CD3 CD4 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD8 CD1a R4, T6, Leu6, HTA1 b-2-Microglobulin, CD74 + + + – + – – – CD74 DHLAG, HLADG, Ia-g, li, invariant chain HLA-DR, CD44 + + + + + + CD158g KIR2DS5 + + CD248 TEM1, Endosialin, CD164L1, MGC119478, MGC119479 Collagen I/IV Fibronectin + ST6GAL1, MGC48859, SIAT1, ST6GALL, ST6N, ST6 b-Galactosamide a-2,6-sialyl- CD1b R1, T6m Leu6 b-2-Microglobulin + + + – + – – – CD75 CD22 CD158h KIR2DS1, p50.1 HLA-C + + CD249 APA, gp160, EAP, ENPEP + + tranferase, Sialo-masked lactosamine, Carbohydrate of a2,6 sialyltransferase + – – + – – + – – CD1c M241, R7, T6, Leu6, BDCA1 b-2-Microglobulin + + + – + – – – CD75S a2,6 Sialylated lactosamine CD22 (proposed) + + – – + + – + + + CD158i KIR2DS4, p50.3 HLA-C + – + CD252 TNFSF4, -

Review the Role of Junctional Adhesion Molecules in Cell-Cell
Histol Histopathol (2005) 20: 197-203 Histology and http://www.hh.um.es Histopathology Cellular and Molecular Biology Review The role of junctional adhesion molecules in cell-cell interactions T. Keiper1, S. Santoso2, P.P. Nawroth1, V. Orlova1 and T. Chavakis1 1Department of Internal Medicine I, University Heidelberg, Heidelberg and 2Institute for Clinical Immunology and Transfusion Medicine, Justus-Liebig-University, Giessen, Germany Summary. Cell-cell-interactions are important for the intracellularly to cytoskelettal signaling molecules such regulation of tissue integrity, the generation of barriers as zonula occludens-1 (ZO-1) or cingulin. (ii) Adherens between different tissues and body compartments junctions (zonula adherens) formed by cadherins, i.e. thereby providing an effective defence against toxic or transmembrane glycoproteins that promote calcium- pathogenic agents, as well as for the regulation of dependent, homophilic cell-cell contacts. The link inflammatory cell recruitment. Intercellular interactions between cell-membrane-associated cadherins and the are regulated by adhesion receptors on adjacent cells actin cytoskeleton is mediated by intracellular catenins. which upon extracellular ligand binding mediate VE-cadherin is a member of this family specifically intracellular signals. In the vasculature, neighbouring found in the inter-endothelial junctions. (iii) Gap endothelial cells interact with each other through various junctions are clusters of transmembrane hydrophilic adhesion molecules leading to the generation of channels that do not serve a cellular contact function, but junctional complexes like tight junctions (TJs) and promote the exchange of ions and small molecules adherens junctions (AJs) which regulate both leukocyte relevant for lateral transmission of molecular endothelial interactions and paracellular permeability. In information. Here, we will focus on the family of JAMs, this context, emerging evidence points to the importance which are components of the tight junctions. -

Supplement, Table 12
Supplement, Table 12. Genes affected by Celecoxib in "cell adhesion" Gene Ontology category Geometri UniGene HUGO Parametric Gene c mean of Cluster ID Symbol p-value ratios P/F Hs.93029 SPOCK sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 0.623 0.00192 Hs.166011 CTNND1 catenin (cadherin-associated protein), delta 1 0.665 0.00006 Hs.334131 CDH2 cadherin 2, type 1, N-cadherin (neuronal) 0.677 0.00280 Hs.423 PAP pancreatitis-associated protein 0.683 0.00400 Hs.85266 ITGB4 integrin, beta 4 0.727 0.02171 Hs.407546 TNFAIP6 tumor necrosis factor, alpha-induced protein 6 0.736 0.00532 Hs.432855 LAMC1 laminin, gamma 1 (formerly LAMB2) 0.751 0.00334 Hs.436593 LMAN1 lectin, mannose-binding, 1 0.754 0.00156 Hs.222 ITGA9 integrin, alpha 9 0.759 0.00809 Hs.8037 TM4SF9 transmembrane 4 superfamily member 9 0.759 0.00146 Hs.25051 PKP2 plakophilin 2 0.774 0.07266 Hs.89436 CDH17 cadherin 17, LI cadherin (liver-intestine) 0.774 0.00173 Hs.48998 FLRT2 fibronectin leucine rich transmembrane protein 2 0.776 0.04264 Hs.173548 NRP1 neuropilin 1 0.784 0.00752 protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting Hs.128312 PPFIA1 protein (liprin), alpha 1 0.785 0.00002 Hs.436042 CXCL12 chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) 0.788 0.01889 Hs.409546 ARHGAP5 Rho GTPase activating protein 5 0.789 0.01338 Hs.445120 LAMA2 laminin, alpha 2 (merosin, congenital muscular dystrophy) 0.792 0.00052 Hs.436494 JAM2 junctional adhesion molecule 2 0.81 0.01923 Hs.296049 MFAP4 microfibrillar-associated