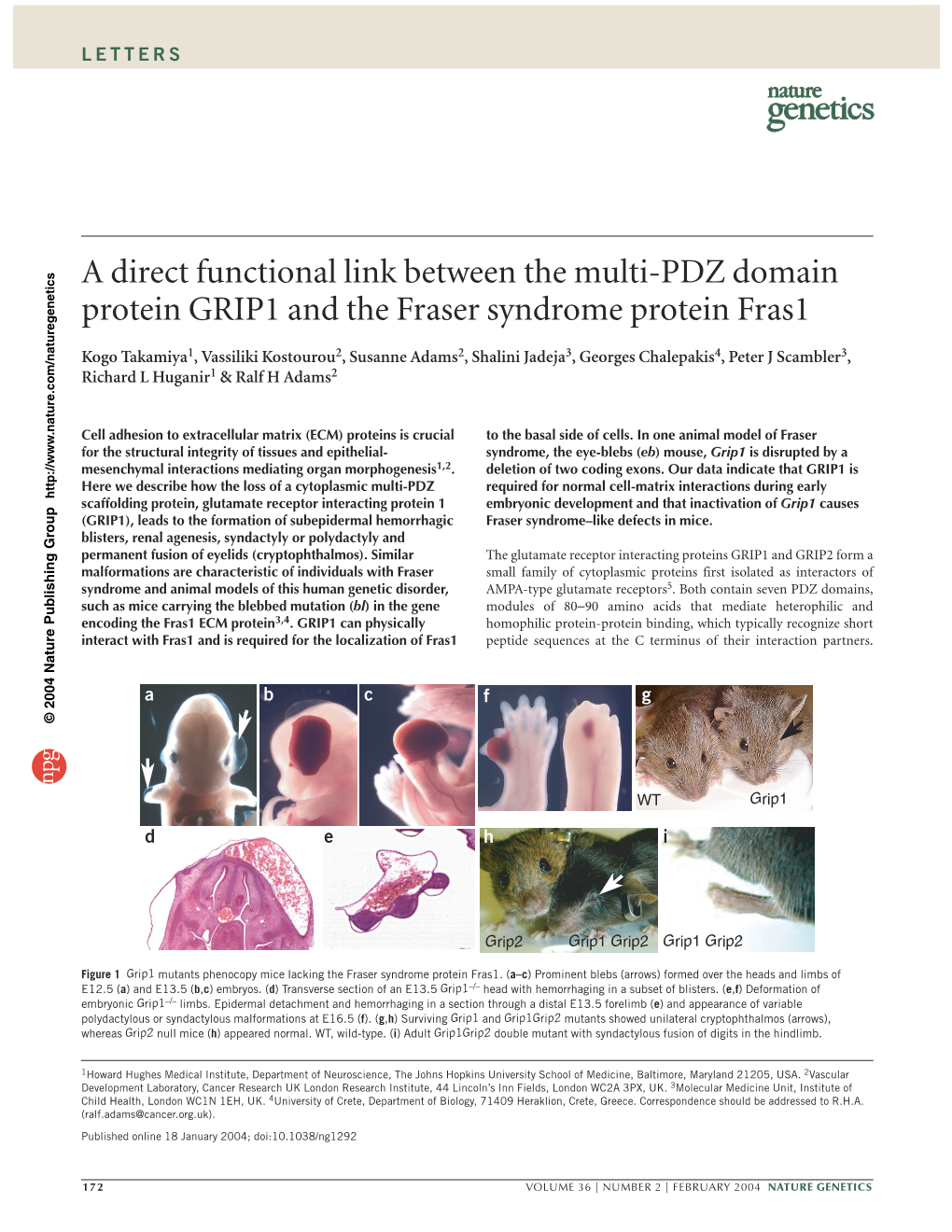
A Direct Functional Link Between the Multi-PDZ Domain Protein GRIP1 and the Fraser Syndrome Protein Fras1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Variants of Innate CD8+ T Cells Are Associated with Grip2 and Klf15 Genes
Cellular & Molecular Immunology www.nature.com/cmi CORRESPONDENCE Variants of innate CD8+ T cells are associated with Grip2 and Klf15 genes Yuna Jo1, Lois Balmer 2,3, Byunghyuk Lee1, Ju A Shim1, Laraib Amir Ali1, Grant Morahan2 and Changwan Hong 1 Cellular & Molecular Immunology (2020) 17:1007–1009; https://doi.org/10.1038/s41423-019-0357-3 Lineage commitment of thymocytes can be distinguished and Next, the frequency of i8Ts was compared with the genotypes modulated by key factors of transcriptional networks, such as of each CC and founder strain. Mapping was performed using the antagonistic expression and function of Th-POK and a logistic regression model (LRM) fit over the reconstructed Runx3 in the CD4 and CD8 lineages1 and Foxp32 and PLZF3 in haplotype matrix.8 The resulting genome-wide distribution of –log CD4+ regulatory T (Treg) cells and natural killer T (NKT) cells, (P) values (ANOVA chi-squared) is shown in Fig. S2a, together with respectively. Little is known, however, about the transcriptional the FDR thresholds. The genome-wide scan for genetic loci regulation of the innate T cell lineage, especially innate CD8+ associated with changes in i8Ts revealed a major peak on T cells (i8Ts), which were initially defined in genetically chromosome 6, centered at 93.7 Mb (Fig. S2b). Within the LOD-2 manipulated mice4 and later in BALB/c mice5 and humans.6 region (89.4–95.6 Mb), Klf15 was the only gene that had a i8Ts are nonconventional αβT cells that exhibit features of nonsynonymous SNP in only the WSB and CAST strains, which memory phenotype and are dependent on IL-4 produced by had the highest i8T levels. -

Molecular Characterisation of Renal Cell Carcinoma and Related Disorders
MOLECULAR CHARACTERISATION OF RENAL CELL CARCINOMA AND RELATED DISORDERS by MARIAM JAFRI A thesis submitted to the University of Birmingham for the degree of DOCTOR OF PHILOSOPHY School of Clinical and Experimental Medicine College of Medical and Dentistry September 2015 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. ABSTRACT Over the last two decades genetic advances have provided novel insights into the molecular basis of familial and sporadic cancers and provided the basis for the development of novel therapeutic approaches. For example, the identification of the gene for von Hippel Lindau disease provided seminal insights into its role in most clear cell renal carcinomas (RCC) and led to new treatments for RCC. In this thesis I investigated three related genetic aspects of neoplasia. Firstly, I analyzed the results of genetic testing for inherited phaeochromocytoma and investigated how clinical features could be used to stratify patients and improve the cost effectiveness of genetic testing. Secondly, I sought to identify novel causes of inherited neoplasia. Through exome sequencing of familial RCC kindreds, CDKN2B was identified as a novel familial RCC gene. -

UC San Diego Electronic Theses and Dissertations
UC San Diego UC San Diego Electronic Theses and Dissertations Title Cardiac Stretch-Induced Transcriptomic Changes are Axis-Dependent Permalink https://escholarship.org/uc/item/7m04f0b0 Author Buchholz, Kyle Stephen Publication Date 2016 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO Cardiac Stretch-Induced Transcriptomic Changes are Axis-Dependent A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Bioengineering by Kyle Stephen Buchholz Committee in Charge: Professor Jeffrey Omens, Chair Professor Andrew McCulloch, Co-Chair Professor Ju Chen Professor Karen Christman Professor Robert Ross Professor Alexander Zambon 2016 Copyright Kyle Stephen Buchholz, 2016 All rights reserved Signature Page The Dissertation of Kyle Stephen Buchholz is approved and it is acceptable in quality and form for publication on microfilm and electronically: Co-Chair Chair University of California, San Diego 2016 iii Dedication To my beautiful wife, Rhia. iv Table of Contents Signature Page ................................................................................................................... iii Dedication .......................................................................................................................... iv Table of Contents ................................................................................................................ v List of Figures ................................................................................................................... -

Mechanism-Driven Hypothesis Generation Support for a Predictive Adverse Effect in Colorectal Cancer Treatment
Aus dem Institut für Medizinische Informationsverarbeitung, Biometrie und Epidemiologie, Lehrstuhl Biometrie und Bioinformatik, Ludwig-Maximilians-Universität München Vorstand: Prof. Dr. Ulrich Mansmann Mechanism-driven Hypothesis Generation Support for a Predictive Adverse Effect in Colorectal Cancer Treatment Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften an der Medizinischen Fakultät der Ludwig-Maximilians-Universität zu München vorgelegt von Sebastian Schaaf aus Bergisch Gladbach 2018 ______________________________________________________ Mit Genehmigung der Medizinischen Fakultät der Universität München Betreuer: Prof. Dr. Ulrich Mansmann Zweitgutachter: Prof. Dr. Volker Heun Dekan: Prof. Dr. med. dent. Reinhard Hickel Tag der mündlichen Prüfung: 15.02.2019 Eidesstattliche Versicherung Schaaf, Sebastian Name, Vorname Ich erkläre hiermit an Eides statt, dass ich die vorliegende Dissertation mit dem Thema „Mechanism-driven Hypothesis Generation Support for a Predictive Adverse Effect in Colorectal Cancer Treatment“ selbstständig verfasst, mich außer der angegebenen keiner weiteren Hilfsmittel bedient und alle Erkenntnisse, die aus dem Schrifttum ganz oder annähernd übernommen sind, als solche kenntlich gemacht und nach ihrer Herkunft unter Bezeichnung der Fundstelle einzeln nachgewiesen habe. Ich erkläre des Weiteren, dass die hier vorgelegte Dissertation nicht in gleicher oder in ähnlicher Form bei einer anderen Stelle zur Erlangung eines akademischen Grades eingereicht wurde. Kerpen, 15.01.2020___________ Sebastian_Schaaf_________________ -

The Role of Lamin Associated Domains in Global Chromatin Organization and Nuclear Architecture
THE ROLE OF LAMIN ASSOCIATED DOMAINS IN GLOBAL CHROMATIN ORGANIZATION AND NUCLEAR ARCHITECTURE By Teresa Romeo Luperchio A dissertation submitted to The Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland March 2016 © 2016 Teresa Romeo Luperchio All Rights Reserved ABSTRACT Nuclear structure and scaffolding have been implicated in expression and regulation of the genome (Elcock and Bridger 2010; Fedorova and Zink 2008; Ferrai et al. 2010; Li and Reinberg 2011; Austin and Bellini 2010). Discrete domains of chromatin exist within the nuclear volume, and are suggested to be organized by patterns of gene activity (Zhao, Bodnar, and Spector 2009). The nuclear periphery, which consists of the inner nuclear membrane and associated proteins, forms a sub- nuclear compartment that is mostly associated with transcriptionally repressed chromatin and low gene expression (Guelen et al. 2008). Previous studies from our lab and others have shown that repositioning genes to the nuclear periphery is sufficient to induce transcriptional repression (K L Reddy et al. 2008; Finlan et al. 2008). In addition, a number of studies have provided evidence that many tissue types, including muscle, brain and blood, use the nuclear periphery as a compartment during development to regulate expression of lineage specific genes (Meister et al. 2010; Szczerbal, Foster, and Bridger 2009; Yao et al. 2011; Kosak et al. 2002; Peric-Hupkes et al. 2010). These large regions of chromatin that come in molecular contact with the nuclear periphery are called Lamin Associated Domains (LADs). The studies described in this dissertation have furthered our understanding of maintenance and establishment of LADs as well as the relationship of LADs with the epigenome and other factors that influence three-dimensional chromatin structure. -

A Meta-Analysis of the Effects of High-LET Ionizing Radiations in Human Gene Expression
Supplementary Materials A Meta-Analysis of the Effects of High-LET Ionizing Radiations in Human Gene Expression Table S1. Statistically significant DEGs (Adj. p-value < 0.01) derived from meta-analysis for samples irradiated with high doses of HZE particles, collected 6-24 h post-IR not common with any other meta- analysis group. This meta-analysis group consists of 3 DEG lists obtained from DGEA, using a total of 11 control and 11 irradiated samples [Data Series: E-MTAB-5761 and E-MTAB-5754]. Ensembl ID Gene Symbol Gene Description Up-Regulated Genes ↑ (2425) ENSG00000000938 FGR FGR proto-oncogene, Src family tyrosine kinase ENSG00000001036 FUCA2 alpha-L-fucosidase 2 ENSG00000001084 GCLC glutamate-cysteine ligase catalytic subunit ENSG00000001631 KRIT1 KRIT1 ankyrin repeat containing ENSG00000002079 MYH16 myosin heavy chain 16 pseudogene ENSG00000002587 HS3ST1 heparan sulfate-glucosamine 3-sulfotransferase 1 ENSG00000003056 M6PR mannose-6-phosphate receptor, cation dependent ENSG00000004059 ARF5 ADP ribosylation factor 5 ENSG00000004777 ARHGAP33 Rho GTPase activating protein 33 ENSG00000004799 PDK4 pyruvate dehydrogenase kinase 4 ENSG00000004848 ARX aristaless related homeobox ENSG00000005022 SLC25A5 solute carrier family 25 member 5 ENSG00000005108 THSD7A thrombospondin type 1 domain containing 7A ENSG00000005194 CIAPIN1 cytokine induced apoptosis inhibitor 1 ENSG00000005381 MPO myeloperoxidase ENSG00000005486 RHBDD2 rhomboid domain containing 2 ENSG00000005884 ITGA3 integrin subunit alpha 3 ENSG00000006016 CRLF1 cytokine receptor like -

Effective Detection of Rare Variants in Pooled DNA Samples Using Cross
Niranjan et al. Genome Biology 2011, 12:R93 http://genomebiology.com/2011/12/9/R93 METHOD Open Access Effective detection of rare variants in pooled DNA samples using Cross-pool tailcurve analysis Tejasvi S Niranjan1,2†, Abby Adamczyk1†, Héctor Corrada Bravo3,4†, Margaret A Taub5, Sarah J Wheelan5,6, Rafael Irizarry5 and Tao Wang1* Abstract Sequencing targeted DNA regions in large samples is necessary to discover the full spectrum of rare variants. We report an effective Illumina sequencing strategy utilizing pooled samples with novel quality (Srfim) and filtering (SERVIC4E) algorithms. We sequenced 24 exons in two cohorts of 480 samples each, identifying 47 coding variants, including 30 present once per cohort. Validation by Sanger sequencing revealed an excellent combination of sensitivity and specificity for variant detection in pooled samples of both cohorts as compared to publicly available algorithms. Background templates for target DNA regions in large numbers of sam- Next-generation sequencing and computational genomic ples is laborious and costly. Fifth, while pooling samples tools permit rapid, deep sequencing for hundreds to thou- can reduce both labor and costs, it reduces sensitivity for sands of samples [1-3]. Recently, rare variants of large the identification of rare variants using currently available effect have been recognized as conferring substantial risks next-generation sequencing strategies and bioinformatics for common diseases and complex traits in humans [4]. tools [1,3]. There is considerable interest in sequencing limited geno- We have optimized a flexible and efficient strategy that mic regions such as sets of candidate genes and target combines a PCR-based amplicon ligation method for regions identified by linkage and/or association studies. -

CM1-Driven Assembly and Activation of Yeast G-Tubulin Small Complex
RESEARCH ARTICLE CM1-driven assembly and activation of yeast g-tubulin small complex underlies microtubule nucleation Axel F Brilot1, Andrew S Lyon1†‡, Alex Zelter2, Shruthi Viswanath3§, Alison Maxwell1, Michael J MacCoss4, Eric G Muller2, Andrej Sali3, Trisha N Davis2, David A Agard1* 1Department of Biochemistry and Biophysics, University of California at San Francisco, San Francisco, United States; 2Department of Biochemistry, University of Washington, Seattle, United States; 3Department of Bioengineering and Therapeutic Sciences, University of California at San Francisco, San Francisco, United States; 4Department of Genome Sciences, University of Washington, Seattle, United States *For correspondence: [email protected] Abstract Microtubule (MT) nucleation is regulated by the g-tubulin ring complex (gTuRC), Present address: †Department conserved from yeast to humans. In Saccharomyces cerevisiae, gTuRC is composed of seven of Biophysics and Howard identical g-tubulin small complex (gTuSC) sub-assemblies, which associate helically to template MT Hughes Medical Institute, growth. gTuRC assembly provides a key point of regulation for the MT cytoskeleton. Here, we University of Texas Southwestern combine crosslinking mass spectrometry, X-ray crystallography, and cryo-EM structures of both Medical Center, Dallas, United monomeric and dimeric gTuSCs, and open and closed helical gTuRC assemblies in complex with ‡ States; Howard Hughes Medical Spc110p to elucidate the mechanisms of gTuRC assembly. gTuRC assembly is substantially aided by Institute, University of Texas the evolutionarily conserved CM1 motif in Spc110p spanning a pair of adjacent gTuSCs. By Southwestern Medical Center, providing the highest resolution and most complete views of any gTuSC assembly, our structures Dallas, United States; § National allow phosphorylation sites to be mapped, surprisingly suggesting that they are mostly inhibitory. -

Systematic Detection of Brain Protein-Coding Genes Under Positive Selection During Primate Evolution and Their Roles in Cognition
Downloaded from genome.cshlp.org on October 7, 2021 - Published by Cold Spring Harbor Laboratory Press Title: Systematic detection of brain protein-coding genes under positive selection during primate evolution and their roles in cognition Short title: Evolution of brain protein-coding genes in humans Guillaume Dumasa,b, Simon Malesysa, and Thomas Bourgerona a Human Genetics and Cognitive Functions, Institut Pasteur, UMR3571 CNRS, Université de Paris, Paris, (75015) France b Department of Psychiatry, Université de Montreal, CHU Ste Justine Hospital, Montreal, QC, Canada. Corresponding author: Guillaume Dumas Human Genetics and Cognitive Functions Institut Pasteur 75015 Paris, France Phone: +33 6 28 25 56 65 [email protected] Dumas, Malesys, and Bourgeron 1 of 40 Downloaded from genome.cshlp.org on October 7, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract The human brain differs from that of other primates, but the genetic basis of these differences remains unclear. We investigated the evolutionary pressures acting on almost all human protein-coding genes (N=11,667; 1:1 orthologs in primates) based on their divergence from those of early hominins, such as Neanderthals, and non-human primates. We confirm that genes encoding brain-related proteins are among the most strongly conserved protein-coding genes in the human genome. Combining our evolutionary pressure metrics for the protein- coding genome with recent datasets, we found that this conservation applied to genes functionally associated with the synapse and expressed in brain structures such as the prefrontal cortex and the cerebellum. Conversely, several genes presenting signatures commonly associated with positive selection appear as causing brain diseases or conditions, such as micro/macrocephaly, Joubert syndrome, dyslexia, and autism. -

Distinct Transcriptomes Define Rostral and Caudal 5Ht Neurons
DISTINCT TRANSCRIPTOMES DEFINE ROSTRAL AND CAUDAL 5HT NEURONS by CHRISTI JANE WYLIE Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Dissertation Advisor: Dr. Evan S. Deneris Department of Neurosciences CASE WESTERN RESERVE UNIVERSITY May, 2010 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of ______________________________________________________ candidate for the ________________________________degree *. (signed)_______________________________________________ (chair of the committee) ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. TABLE OF CONTENTS TABLE OF CONTENTS ....................................................................................... iii LIST OF TABLES AND FIGURES ........................................................................ v ABSTRACT ..........................................................................................................vii CHAPTER 1 INTRODUCTION ............................................................................................... 1 I. Serotonin (5-hydroxytryptamine, 5HT) ....................................................... 1 A. Discovery.............................................................................................. -

Mouse Grip2 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Grip2 Knockout Project (CRISPR/Cas9) Objective: To create a Grip2 knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Grip2 gene (NCBI Reference Sequence: NM_001159507 ; Ensembl: ENSMUSG00000030098 ) is located on Mouse chromosome 6. 24 exons are identified, with the ATG start codon in exon 1 and the TGA stop codon in exon 24 (Transcript: ENSMUST00000162300). Exon 2~12 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Homozygous null mice are born in numbers expected by the Mendelian ratio and show no overt phenotype. Exon 2 starts from about 1.79% of the coding region. Exon 2~12 covers 46.19% of the coding region. The size of effective KO region: ~8114 bp. The KO region does not have any other known gene. Page 1 of 9 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 2 3 4 5 6 7 8 9 10 11 12 24 Legends Exon of mouse Grip2 Knockout region Page 2 of 9 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section upstream of Exon 2 is aligned with itself to determine if there are tandem repeats. Tandem repeats are found in the dot plot matrix. The gRNA site is selected outside of these tandem repeats. Overview of the Dot Plot (down) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 361 bp section downstream of Exon 12 is aligned with itself to determine if there are tandem repeats. -
A Glutamate Receptor–Interacting Protein Homolog Organizes Muscle Guidance in Drosophila
Downloaded from genesdev.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press A Glutamate Receptor–Interacting Protein homolog organizes muscle guidance in Drosophila Laura E. Swan,1,5 Carolin Wichmann,1,5 Ulrike Prange,1 Andreas Schmid,1 Manuela Schmidt,1 Tobias Schwarz,2 Evgeni Ponimaskin,2 Frank Madeo,3 Gerd Vorbru¨ggen,4 and Stephan J. Sigrist1,6 1European Neuroscience Institute Go¨ttingen, Max-Planck-Society, 37073 Go¨ttingen, Germany; 2Department of Neuro and Sensory Physiology, Medical School, University of Go¨ttingen, 37073 Go¨ttingen, Germany; 3Institute for Physiological Chemistry, University of Tu¨bingen, 72076 Tu¨bingen, Germany; 4Max-Planck-Institute for Biophysical Chemistry, 37077 Go¨ttingen, Germany During Drosophila embryogenesis, developing muscles extend growth-cone–like structures to navigate toward specific epidermal attachment sites. Here, we show that the homolog of Glutamate Receptor–Interacting Proteins (DGrip) acts as a key component of proper muscle guidance. Mutations in dgrip impair patterning of ventral longitudinal muscles (VLMs), whereas lateral transverse muscles (LTMs) that attach to intrasegmental attachment sites develop normally. Myoblast fusion, stabilization of muscle contacts, and general muscle function are not impaired in the absence of DGrip. Instead, the proper formation of cellular extensions during guidance fails in dgrip mutant VLMs. DGrip protein concentrates at the ends of VLMs while these muscles guide toward segment border attachment sites. Conversely, LTMs overexpressing DGrip form ectopic cellular extensions that can cause attachment of these muscles to other muscles at segment borders. Our data suggest that DGrip participates in the reception of an attractive signal that emanates from the epidermal attachment sites to direct the motility of developing muscles.