Pathogen and Host Genetics Underpinning Cryptococcal Disease
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Diversity of the Cryptococcus Neoformans-Cryptococcus Gattii Species Complex Marjan Bovers1, Ferry Hagen1 and Teun Boekhout1,2
S4 Rev Iberoam Micol 2008; 25: S4-S12 Review Diversity of the Cryptococcus neoformans-Cryptococcus gattii species complex Marjan Bovers1, Ferry Hagen1 and Teun Boekhout1,2 1Yeast Research Group, CBS Fungal Diversity Centre, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands; 2Department of Internal Medicine and Infectious Diseases, University Medical Centre Utrecht, The Netherlands Summary More than 110 years of study of the Cryptococcus neoformans and Cryptococcus gattii species complex has resulted in an enormous accumulation of fundamental, applied biological and clinical knowledge. Recent developments in our understanding of the diversity within the species complex are presented, emphasizing the intraspecific complexity, which includes species, microspecies, hybrids, serotypes and genotypes. Each of these may have different roles in disease. An overview of obsolete and current names is presented. Key words Cryptococcus neoformans, Cryptococcus gattii, Yeast, Taxonomy, Pathogen. Diversidad del complejo de especies Cryptococcus neoformans-Cryptococcus gattii Resumen Más de 110 años de estudio sobre el complejo de especies Cryptococcus neoformans y Cryptococcus gattii han dado lugar a un gran acúmulo de conocimiento básico, aplicado y clínico. En este artículo se describen los avances recientes en la comprensión de su diversidad, haciendo énfasis en la complejidad intraespecífica que presenta y que llega a englobar especies, microespecies, híbridos, serotipos y genotipos. Cada uno de estos grupos puede jugar un papel diferente en la enfermedad. Se presenta también una visión global de los nombres obsoletos y actuales de estos taxones. Palabras clave Cryptococcus neoformans, Cryptococcus gattii, Levaduras, Patógeno, Taxonomía. Introduction racteristics of the genus Saccharomyces were not present, he placed these species in the genus Cryptococcus [166]. -

Cryptococcus Gattii Outbreak
The recent Cryptococcus gattii outbreak A deadly pathway… From Trees to Lungs to Brain ryptococcus gattii disease. The route of C has been in the infection is from breathing the news lately, due to a recent airborne organism, which outbreak in the Pacific becomes lodged in the Northwest. This new, more pulmonary tissues. virulent strain is expected to by Jay Hardy, CLS, SM (NRCM) spread further throughout the Symptoms Northwest and Northern California in the coming The pulmonary disease, Jay Hardy is the founder and months. known as cpryptococcosis, president of Hardy Diagnostics. develops slowly. After He began his career in exposure it can take two to microbiology as a Medical Technologist in Santa Barbara, twelve months for symptoms California. to appear, with the usual onset In 1980, he began at six or seven months. manufacturing culture media for the local hospitals. Today, The symptoms include: Hardy Diagnostics is the third largest media manufacturer in the U.S. • Cough that lasts weeks or months To ensure rapid and reliable turn around time, Hardy • Sharp chest pain Figure 1: An India ink stain Diagnostics maintains six • Unexplained shortness of showing the capsule surrounding distribution centers, and breath produces over 3,000 products the C. gattii cells in the yeast form used in clinical and industrial at 1,000X. Photo from Haley/CDC. • Severe headache microbiology laboratories • Confusion throughout the world. The organism is a fungus that • Fever is usually found in the soil • Night sweats www.HardyDiagnostics.com and on trees. It is not spread • Unintended weight loss from human to human or animal to human. -

A Silver Bullet in a Golden Age of Functional Genomics: the Impact of Agrobacterium-Mediated Transformation of Fungi
Idnurm, A. , Bailey, A. M., Cairns, T., Elliott, C., Foster, G., Ianiri, G., & Jeon, J. (2017). A silver bullet in a golden age of functional genomics: the impact of Agrobacterium-mediated transformation of fungi. Fungal Biology and Biotechnology, 4, [4:6]. https://doi.org/10.1186/s40694- 017-0035-0 Publisher's PDF, also known as Version of record License (if available): CC BY Link to published version (if available): 10.1186/s40694-017-0035-0 Link to publication record in Explore Bristol Research PDF-document This is the final published version of the article (version of record). It first appeared online via BMC at https://fungalbiolbiotech.biomedcentral.com/articles/10.1186/s40694-017-0035-0. Please refer to any applicable terms of use of the publisher. University of Bristol - Explore Bristol Research General rights This document is made available in accordance with publisher policies. Please cite only the published version using the reference above. Full terms of use are available: http://www.bristol.ac.uk/red/research-policy/pure/user-guides/ebr-terms/ Idnurm et al. Fungal Biol Biotechnol (2017) 4:6 DOI 10.1186/s40694-017-0035-0 Fungal Biology and Biotechnology REVIEW Open Access A silver bullet in a golden age of functional genomics: the impact of Agrobacterium‑mediated transformation of fungi Alexander Idnurm1* , Andy M. Bailey2, Timothy C. Cairns3, Candace E. Elliott1, Gary D. Foster2, Giuseppe Ianiri4 and Junhyun Jeon5 Abstract The implementation of Agrobacterium tumefaciens as a transformation tool revolutionized approaches to discover and understand gene functions in a large number of fungal species. A. -

Associations Between Cryptococcus Genotypes, Phenotypes, and Clinical Parameters of Human Disease: a Review
Journal of Fungi Review Associations between Cryptococcus Genotypes, Phenotypes, and Clinical Parameters of Human Disease: A Review Marhiah C. Montoya 1,2 , Paul M. Magwene 3 and John R. Perfect 1,2,* 1 Division of Infectious Diseases, Department of Medicine, Duke University, Durham, NC 27710, USA; [email protected] 2 Division of Infectious Diseases, Department of Molecular Genetics and Microbiology, Duke University, Durham, NC 27710, USA 3 Department of Biology, Duke University, Durham, NC 27710, USA; [email protected] * Correspondence: [email protected] Abstract: The genus Cryptococcus contains two primary species complexes that are significant op- portunistic human fungal pathogens: C. neoformans and C. gattii. In humans, cryptococcosis can manifest in many ways, but most often results in either pulmonary or central nervous system disease. Patients with cryptococcosis can display a variety of symptoms on a spectrum of severity because of the interaction between yeast and host. The bulk of our knowledge regarding Cryptococcus and the mechanisms of disease stem from in vitro experiments and in vivo animal models that make a fair attempt, but do not recapitulate the conditions inside the human host. To better understand the dynamics of initiation and progression in cryptococcal disease, it is important to study the genetic and phenotypic differences in the context of human infection to identify the human and fungal risk factors that contribute to pathogenesis and poor clinical outcomes. In this review, we summarize the current understanding of the different clinical presentations and health outcomes that are associated with pathogenicity and virulence of cryptococcal strains with respect to specific Citation: Montoya, M.C.; Magwene, P.M.; Perfect, J.R. -

Yeasts Diversity in Brazilian Cerrado Soils: Study of the Enzymatic Activities
Vol. 7(32), pp. 4176-4190, 9 August, 2013 DOI: 10.5897/AJMR2013.5752 ISSN 1996-0808 ©2013 Academic Journals African Journal of Microbiology Research http://www.academicjournals.org/AJMR Full Length Research Paper Yeasts diversity in Brazilian Cerrado soils: Study of the enzymatic activities Fernanda Paula Carvalho1, Angélica Cristina de Souza1, Karina Teixeira Magalhães-Guedes1*, Disney Ribeiro Dias2, Cristina Ferreira Silva1 and Rosane Freitas Schwan1 1Department of Biology, Federal University of Lavras (UFLA), Campus Universitário, 37.200-000, Lavras, MG, Brazil. 2Department of Food Science, Federal University of Lavras (UFLA), Campus Universitário, 37.200-000, Lavras, MG, Brazil. Accepted 22 July, 2013 A total of 307 yeasts strains were isolated from native Cerrado (Brazilian Savannah) soils collected at Passos, Luminárias and Arcos areas in Minas Gerais State, Brazil. The soils were chemically characterized. Ten yeast genera (Candida, Cryptococcus, Debaryomyces, Kazachstania, Kodamaea, Lindnera, Pichia, Schwanniomyces, Torulaspora and Trichosporon) and 23 species in both rainy and dry seasons were identified. All genera were abundant during the dry season. The pH values of the soil from the Passos, Luminárias and Arcos areas varied from 4.1 to 5.5. There were no significant differences in the concentrations of phosphorus, magnesium and organic matter in the soils among the studied areas. The Arcos area contained large amounts of aluminum during the rainy season and both hydrogen and aluminum in the rainy and dry seasons. The yeast populations identified seemed to be unaffected by the high levels of aluminum in the soil. The API ZYM® (BioMérieux, France) system was employed to characterize the extracellular enzymatic activity profiles of the yeast isolates. -

Health and Disease Status in a Threatened Marsupial, the Quokka (Setonix Brachyurus)
Health and disease status in a threatened marsupial, the quokka (Setonix brachyurus) Pedro Martínez-Pérez Veterinarian (Hons), MVS Conservation Medicine This thesis was submitted in fulfilment of the requirements for the degree of Doctor of Philosophy in Wildlife Veterinary Studies, Murdoch University School of Veterinary and Life Sciences Murdoch University Murdoch, Western Australia January 2016 Declaration I declare that this thesis is my own account of my research and contains as its main content, work which has not been previously submitted for a degree at any tertiary educational institution. _______________________________________________ Pedro A. Martínez-Pérez I Abstract Between 1901 and 1931, there were at least six anecdotal records of disease outbreaks in mainland quokkas (Setonix brachyurus) that were associated with mass. This time period pre-dates the arrival of the red fox (Vulpes vulpes). Despite these outbreaks, little or no research has been carried out to establish health and disease baseline data of the fragmented and scattered, extant populations. Epidemiological data was determined for a range of potential pathogens, and established physiological reference intervals of apparently healthy, wild quokkas on Rottnest Island and mainland locations. There were significant differences between Rottnest Island and mainland quokkas. Rottnest Island animals had haemograms with mark evidence of oxidative injury and bone marrow response consistent with a regenerative normocytic hypochromic anaemia. Except alkaline phosphatase (ALP), all blood chemistry analytes where higher in mainland animals, with particular emphasis on creatine kinase (CK), alanine amino transferase (ALT), aspartate amino transferase (AST) and vitamin E. Some other key findings include a widespread presence of a novel herpesvirus (MaHV-6), the recovery of Cryptococcus neoformans var. -

A Silver Bullet in a Golden Age of Functional Genomics: the Impact of Agrobacterium-Mediated Transformation of Fungi
Idnurm et al. Fungal Biol Biotechnol (2017) 4:6 DOI 10.1186/s40694-017-0035-0 Fungal Biology and Biotechnology REVIEW Open Access A silver bullet in a golden age of functional genomics: the impact of Agrobacterium‑mediated transformation of fungi Alexander Idnurm1* , Andy M. Bailey2, Timothy C. Cairns3, Candace E. Elliott1, Gary D. Foster2, Giuseppe Ianiri4 and Junhyun Jeon5 Abstract The implementation of Agrobacterium tumefaciens as a transformation tool revolutionized approaches to discover and understand gene functions in a large number of fungal species. A. tumefaciens mediated transformation (AtMT) is one of the most transformative technologies for research on fungi developed in the last 20 years, a development arguably only surpassed by the impact of genomics. AtMT has been widely applied in forward genetics, whereby generation of strain libraries using random T-DNA insertional mutagenesis, combined with phenotypic screening, has enabled the genetic basis of many processes to be elucidated. Alternatively, AtMT has been fundamental for reverse genet- ics, where mutant isolates are generated with targeted gene deletions or disruptions, enabling gene functional roles to be determined. When combined with concomitant advances in genomics, both forward and reverse approaches using AtMT have enabled complex fungal phenotypes to be dissected at the molecular and genetic level. Addition- ally, in several cases AtMT has paved the way for the development of new species to act as models for specifc areas of fungal biology, particularly in plant pathogenic ascomycetes and in a number of basidiomycete species. Despite its impact, the implementation of AtMT has been uneven in the fungi. This review provides insight into the dynamics of expansion of new research tools into a large research community and across multiple organisms. -

Maternal–Fetal Transmission of Cryptococcus Gattii in Harbor Porpoise
LETTERS Oral Oncul, Yunus Atalay, Maternal–Fetal www.cdc.gov/EID/content/17/2/302- Yalcin Onem, Vedat Turhan, Ali appF.htm). The fi rst stomach chamber Acar, Yavuz Uyar, Transmission of contained two 3.5 cm × 2.5 cm raised, Dilek Y. Caglayik, Sezai Ozkan, Cryptococcus gattii centrally umbilicated ulcers and sev- and Levent Gorenek in Harbor Porpoise eral embedded anisakid nematodes. Author affi liations: Gulhane Military Medi- The uterus was gravid in the right cal Academy Haydarpasa Training Hospi- To the Editor: We report mater- horn with a mid-term fetus. No other tal, Istanbul, Turkey (O. Oncul, Y. Atalay, nal–fetal transmission of Cryptococ- gross lesions were identifi ed. Micro- Y. Onem, V. Turhan, A. Acar, S. Ozkan, L. cus gattii and death in a wild porpoise. scopically, the lung lesions correlated Gorenek); and Refi k Saydam National Pub- Cryptococcus neoformans and C. gat- with granulomatous to pyogranuloma- lic Health Agency, Ankara, Turkey (Y. Uyar, tii are 2 environmental, encapsulated tous infi ltrates, often with a myriad of D.Y. Caglayik) yeasts that cause invasive, potentially yeasts. life-threatening infections in humans The male fetus (length 30 cm, DOI: 10.3201/eid1702.100663 and animals (1). C. neoformans causes weight 2.4 kg), was examined sepa- disease in immunocompromised hosts, rately at a different facility than the References and C. gattii is also pathogenic in im- dam. It appeared grossly normal ex- 1. Tang YW, Li YL, Ye KL, Xu ZY, Ruo munocompetent hosts (2). Since 1999, ternally and was at a gestation of ≈5–6 SL, Fisher-Hoch SP, et al. -

Towards an Integrated Phylogenetic Classification of the Tremellomycetes
http://www.diva-portal.org This is the published version of a paper published in Studies in mycology. Citation for the original published paper (version of record): Liu, X., Wang, Q., Göker, M., Groenewald, M., Kachalkin, A. et al. (2016) Towards an integrated phylogenetic classification of the Tremellomycetes. Studies in mycology, 81: 85 http://dx.doi.org/10.1016/j.simyco.2015.12.001 Access to the published version may require subscription. N.B. When citing this work, cite the original published paper. Permanent link to this version: http://urn.kb.se/resolve?urn=urn:nbn:se:nrm:diva-1703 available online at www.studiesinmycology.org STUDIES IN MYCOLOGY 81: 85–147. Towards an integrated phylogenetic classification of the Tremellomycetes X.-Z. Liu1,2, Q.-M. Wang1,2, M. Göker3, M. Groenewald2, A.V. Kachalkin4, H.T. Lumbsch5, A.M. Millanes6, M. Wedin7, A.M. Yurkov3, T. Boekhout1,2,8*, and F.-Y. Bai1,2* 1State Key Laboratory for Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, PR China; 2CBS Fungal Biodiversity Centre (CBS-KNAW), Uppsalalaan 8, Utrecht, The Netherlands; 3Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig 38124, Germany; 4Faculty of Soil Science, Lomonosov Moscow State University, Moscow 119991, Russia; 5Science & Education, The Field Museum, 1400 S. Lake Shore Drive, Chicago, IL 60605, USA; 6Departamento de Biología y Geología, Física y Química Inorganica, Universidad Rey Juan Carlos, E-28933 Mostoles, Spain; 7Department of Botany, Swedish Museum of Natural History, P.O. Box 50007, SE-10405 Stockholm, Sweden; 8Shanghai Key Laboratory of Molecular Medical Mycology, Changzheng Hospital, Second Military Medical University, Shanghai, PR China *Correspondence: F.-Y. -

12 Tremellomycetes and Related Groups
12 Tremellomycetes and Related Groups 1 1 2 1 MICHAEL WEIß ,ROBERT BAUER ,JOSE´ PAULO SAMPAIO ,FRANZ OBERWINKLER CONTENTS I. Introduction I. Introduction ................................ 00 A. Historical Concepts. ................. 00 Tremellomycetes is a fungal group full of con- B. Modern View . ........................... 00 II. Morphology and Anatomy ................. 00 trasts. It includes jelly fungi with conspicuous A. Basidiocarps . ........................... 00 macroscopic basidiomes, such as some species B. Micromorphology . ................. 00 of Tremella, as well as macroscopically invisible C. Ultrastructure. ........................... 00 inhabitants of other fungal fruiting bodies and III. Life Cycles................................... 00 a plethora of species known so far only as A. Dimorphism . ........................... 00 B. Deviance from Dimorphism . ....... 00 asexual yeasts. Tremellomycetes may be benefi- IV. Ecology ...................................... 00 cial to humans, as exemplified by the produc- A. Mycoparasitism. ................. 00 tion of edible Tremella fruiting bodies whose B. Tremellomycetous Yeasts . ....... 00 production increased in China alone from 100 C. Animal and Human Pathogens . ....... 00 MT in 1998 to more than 250,000 MT in 2007 V. Biotechnological Applications ............. 00 VI. Phylogenetic Relationships ................ 00 (Chang and Wasser 2012), or extremely harm- VII. Taxonomy................................... 00 ful, such as the systemic human pathogen Cryp- A. Taxonomy in Flow -
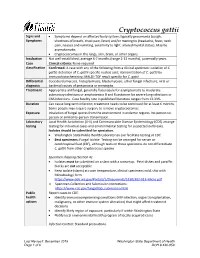
Cryptococcus Gattii Reporting and Investigation Guideline
Cryptococcus gattii Signs and • Symptoms depend on affected body system; typically pneumonia (cough, Symptoms shortness of breath, chest pain, fever) and/or meningitis (headache, fever, neck pain, nausea and vomiting, sensitivity to light, altered mental status). May be asymptomatic. • Cryptococcomas in the lungs, skin, brain, or other organs. Incubation Not well-established; average 6-7 months (range 2-13 months), potentially years. Case Clinical criteria: None required classification Confirmed: A case with any of the following from a clinical specimen: isolation of C. gattii; detection of C. gattii-specific nucleic acid; demonstration of C. gattii by immunohistochemistry; MALDI-TOF result specific for C. gattii Differential Coccidioidomycosis, histoplasmosis, blastomycosis, other fungal infections, viral or diagnosis bacterial causes of pneumonia or meningitis Treatment Appropriate antifungal, generally fluconazole for asymptomatic to moderate pulmonary infections or amphotericin B and flucytosine for severe lung infections or CNS infections. Case fatality rate in published literature ranges from 13-33%. Duration Can cause long-term infection; treatment needs to be continued for at least 6 months. Some people may require surgery to remove cryptococcomas. Exposure Inhalation of fungal spores from the environment in endemic regions. No person-to- person or animal-to-person transmission. Laboratory Local Health Jurisdiction (LHJ) and Communicable Disease Epidemiology (CDE) arrange testing testing for individual cases and environmental testing for suspected outbreaks. Isolates should be submitted for speciation. • Washington State Public Health Laboratories can facilitate testing at CDC • Best specimens: Fungal isolate. Testing can be arranged for serum or cerebrospinal fluid (CSF), although tests on these specimens do not differentiate C. gattii from other Cryptococcus species Specimen shipping (Section 4): • Isolates must be submitted on a slant with a screw top. -

Tremella Mesenterica Xylose 38.00
COMPARATIVE QUALITATIVE ANALYSES OF HYDROLYSIS PRODUCTS OF EXTRACELLULAR POLYSACCHARIDES PRODUCED BY SOME YEASTS AND YEAST-LIKE FUNGI by Patricia E. M. Flodin B.Sc, University of British Columbia, 1969 A THESIS SUBMITTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE in the Department of Botany We accept this thesis as conforming to the required standard THE UNIVERSITY OF BRITISH COLUMBIA August 1972 In presenting this thesis in partial fulfilment of the requirements for an advanced degree at the University of British Columbia, I agree that the Library shall make it freely available for reference and study. I further agree that permission for extensive copying of this thesis for scholarly purposes may be granted by the Head of my Department or by his representatives. It is understood that copying or publication of this thesis for financial gain shall not be allowed without my written permission. Department of The University of British Columbia Vancouver 8, Canada Date ftfy£- /3 } )9 7^L i ABSTRACT The objective of the experiments was to compare ' qualitatively the monosaccharides in the hydrolysis products of the extracellular polysaccharides of several yeasts and yeast-like fungi. Specifically, the study was aimed at finding similarities and differences that might be useful in suggesting and supporting taxonomic relation• ships. Gas chromatography and paper chromatography were used as methods of analyses in an effort to find out what method is sufficient at the qualitative level for distinguishing some genera of yeasts and yeast-like fungi; and what method would be best at the quantitative level for distinguishing amongst some species of the same genus.