Microbial Community Dynamic in Tomato Fruit During Spontaneous Fermentation and Biotechnological Characterization of Indigenous Lactic Acid Bacteria
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Steady State and Transient Behavior of a Continuous Fermentor Thor Almep R Hanson Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1969 Steady state and transient behavior of a continuous fermentor Thor almeP r Hanson Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Chemical Engineering Commons Recommended Citation Hanson, Thor almeP r, "Steady state and transient behavior of a continuous fermentor " (1969). Retrospective Theses and Dissertations. 4109. https://lib.dr.iastate.edu/rtd/4109 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. 70-13,589 HANSONJ Thor Palmer, 1942- STEADY STATE AND TRANSIENT BEHAVIOR OF A CONTINUOUS FERMENTOR. Iowa State University, Ph.D., 1969 Engineering, chemical University Microfilms, Inc., Ann Arbor, Michigan THIS DISSERTATION HAS BEEN MICROFILMED EXACTLY AS RECEIVED STEADY STATE AND TRANSIENT BEHAVIOR OF A CONTINUOUS FERMENTOR Thor Palmer Hanson A Dissertation Submitted to the Graduate Faculty in Partial Fulfillment of The Requirements for the Degree of DOCTOR OP PHILOSOPHY Major Subject: Chemical Engineering Approved : Signature was redacted for privacy. In Charge of Major Work Signature was redacted for privacy. Head of Major Department Signature was redacted for privacy. D( 'ége Iowa State University Ames, lowa 1969 11 TABLE OP CONTENTS Page ABSTRACT v I. INTRODUCTION 1 II. BACKGROUND INFORMATION 5 A. Bacterial Growth 5 B. Mathematical Models for Bacterial Growth l4 1. -

Fermentation Microorganisms and Flavor Changes in Fermented Foods R.F
Fermentation Technology—12th World Congress of Food Science and Technology Fermentation Microorganisms and Flavor Changes in Fermented Foods R.F. MCFEETERS ABSTRACT: Food fermentation processes often result in profound changes in flavor relative to the starting ingredients. However, fermenting foods are typically very complex ecosystems with active enzyme systems from the ingredient materials interacting with the metabolic activities of the fermentation organisms. Factors such as added salt, particle sizes, temperature, and oxygen levels will also have important effects on the chemistry that occurs during fermentation. This is a brief review of recent research on flavor changes in food fermentations. The emphasis will be on the role of lactic acid bacteria in changing the compounds that help determine the character of fermented foods from plant-based substrates. Introduction GC-olfactometry led to recognition of a compound with an odor actic acid bacteria influence the flavor of fermented foods in a close to that of the fermentation brine. The compound with a fer- Lvariety of ways. In many cases, the most obvious change in a lac- mentation brine odor was identified as trans-4-hexenoic acid. They tic acid fermentation is the production of acid and lowering pH that also tentatively identified the presence of cis-4-hexenoic acid. In a results in an increase in sourness. Since most of the acid produced reconstitution experiment, a solution that contained 25 ppm trans- in fermentations will be produced by the metabolism of sugars, 4-hexenoic acid, 10 ppm phenyl ethyl alcohol, 0.65% lactic acid, sweetness will likely decrease as sourness increases. The produc- 0.05% acetic acid, and 8% NaCl had an odor very similar to that of tion of volatile flavor components tends to be the first mechanism brine from fermented cucumbers. -

Effect of Lactic Acid Fermentation on Color, Phenolic Compounds And
microorganisms Article Effect of Lactic Acid Fermentation on Color, Phenolic Compounds and Antioxidant Activity in African Nightshade Alexandre Degrain 1,2, Vimbainashe Manhivi 1, Fabienne Remize 2,* , Cyrielle Garcia 2 and Dharini Sivakumar 1 1 Phytochemical Food Network Research Group, Department of Crop Sciences, Tshwane University of Technology, Pretoria West 0001, South Africa; [email protected] (A.D.); [email protected] (V.M.); [email protected] (D.S.) 2 QualiSud, Université de La Réunion, CIRAD, Université Montpellier, Montpellier SupAgro, Université d’Avignon, 97490 Sainte Clotilde, France; [email protected] * Correspondence: [email protected]; Tel.: +27-012-382-5303 Received: 3 August 2020; Accepted: 23 August 2020; Published: 30 August 2020 Abstract: This study aimed to investigate the influences of fermentation at 37 ◦C for 3 days by different lactic acid bacterium strains, Lactobacillus plantarum (17a), Weissella cibaria (21), Leuconostoc pseudomesenteroides (56), W. cibaria (64) or L. plantarum (75), on color, pH, total soluble solids (TSS), phenolic compounds and antioxidant activity of African nightshade (leaves). Results indicated fermentation with L. plantarum 75 strain significantly decreased the pH and total soluble solids, and increased the concentration of ascorbic acid after 3 days. L. plantarum 75 strain limited the color modification in fermented nightshade leaves and increased the total polyphenol content and the antioxidant activity compared to the raw nightshade leaves. Overall, L. plantarum 75 enhanced the functional potential of nightshade leaves and improved the bioavailability of gallic, vanillic acid, coumaric, ferulic ellagic acids, flavonoids (catechin, quercetin and luteolin) and ascorbic acid compared to the other lactic acid bacterium strains. -
NOTES: Ch 9, Part 4 - 9.5 & 9.6 - Fermentation & Regulation of Cellular Respiration
NOTES: Ch 9, part 4 - 9.5 & 9.6 - Fermentation & Regulation of Cellular Respiration 9.5 - Fermentation enables some cells to produce ATP without the use of oxygen ● Cellular respiration requires O2 to produce ATP ● Glycolysis can produce ATP with or without O2 (in aerobic or anaerobic conditions) ● In the absence of O2, glycolysis couples with fermentation to produce ATP Alternative Metabolic Pathways - Vocabulary: ● aerobic: existing in presence of oxygen ● anaerobic: existing in absence of oxygen ● FERMENTATION = anaerobic catabolism of organic nutrients Types of Fermentation ● Fermentation consists of glycolysis plus reactions that regenerate NAD+, which can be reused by glycolysis ● Two common types are alcohol fermentation and lactic acid fermentation Alcohol Fermentation + Pyruvate + NADH ethanol + CO2 + NAD ● pyruvate is converted to ethanol ● NADH is oxidized to NAD+ (recycled) ● performed by yeast and some bacteria Alcohol Fermentation ● In alcohol fermentation, pyruvate is converted to ethanol in two steps, with the first releasing CO2 ● Alcohol fermentation by yeast is used in brewing, winemaking, and baking 2 ADP + 2 P i 2 ATP Glucose Glycolysis 2 Pyruvate 2 NAD+ 2 NADH 2 CO2 + 2 H+ 2 Ethanol 2 Acetaldehyde Alcohol fermentation Lactic Acid Fermentation Pyruvate + NADH lactic acid + NAD+ ● pyruvate is reduced to lactic acid (3-C compound); no CO2 produced ● NADH is oxidized to NAD+ (recycling of NAD+) Lactic Acid Fermentation ● Lactic acid fermentation by some fungi and bacteria is used to make cheese and yogurt ● Human muscle -

The Living Cell
THE LIVING CELL A Tour of the cell The cell is the smallest and the basic unit of structure of all organisms. There are two main types or categories of cells: prokaryotic cells and eukaryotic cells. Prokaryotic cells are independently functioning organisms, such as microscopic amoeba and bacteria cells whereas, eukaryotic cells generally function only when they are a part of a larger organism such as the cells that make up our body. An exception is yeast which is eukaryotic, yet lives as a single cell. Many different types of cells are found in nature. Cells come in all shapes and sizes and perform various functions. In the body, there are brain cells, skin cells, liver cells, stomach cells and so on. All of these cells have unique functions and features. While typical animal cells are about one-hundredth of a millimeter in diameter (1/100 mm), bacterial cells are only a few hundredth-thousandths of a centimeter in size (1/100,000 cm). Cells also have variety of shapes. Plant cells are often rectangular or polygonal, while egg cells are usually spherical. Bacterial cells may be rod Nerve cells are a complex Muscle cells are extremely shaped or spiral array of fibers elongated a b c Figure 1: a : Rod shaped bacterial cells; b: Nerve cell; c: elongated muscle cell The differences in shapes and sizes reflect the differences in the functions that the cells perform. Elongated muscle cells exert forces when they contract. Branched nerve cells transmit impulses to many other cells. 1 Bacterial cells operate independently and ensure their survival by reproducing in large numbers, whereas animal and plant cells have collective interdependence. -

American Council for Medicinally Active Plants (ACMAP) .10Th Annual and 1St Virtual Conference
Journal of Medicinally Active Plants Volume 10 Issue 2 Vol 10 Issue 2 Supplement 6-30-2021 American Council for Medicinally Active Plants (ACMAP) .10th Annual and 1st Virtual Conference Follow this and additional works at: https://scholarworks.umass.edu/jmap Part of the Plant Sciences Commons Recommended Citation . 2021. "American Council for Medicinally Active Plants (ACMAP) .10th Annual and 1st Virtual Conference." Journal of Medicinally Active Plants 10, (2). https://scholarworks.umass.edu/jmap/vol10/iss2/1 This Article is brought to you for free and open access by ScholarWorks@UMass Amherst. It has been accepted for inclusion in Journal of Medicinally Active Plants by an authorized editor of ScholarWorks@UMass Amherst. For more information, please contact [email protected]. et al.: American Council for Medicinally Active Plants (ACMAP) .10th Annu American Council for Medicinally Active Plants (ACMAP) Leading research on medicinal plants 10TH ANNUAL AND 1ST VIRTUAL C ONFERENCE June 24-25, 2021 Hosted by Rutgers University Visit WWW.ACMAP.ORG For more information and registration Journal of Medicinally Active Plants Vol. 10, Iss. 2 [2021], Welcome back to Rutgers after 10 years: Prof. Jim Simon, 2021 ACMAP Host, Distinguished Professor of Plant Biology, Rutgers University Welcome to School of Biological and Environmental Sciences and Rutgers: Dr. Wendie Cohick, Dean of Research and Graduate Education School of Environmental & Biological Sciences (SEBS), and Director of Research, New Jersey Agricultural Experiment Station (NJAES) Welcome to the ACMAP conference: Dr. Anait S. Levenson, President of ACMAP, Professor of Cancer Research and Pharmacology, Long Island University Jim Simon, Ph.D. Anait S. -

1. Introduction
1 1. INTRODUCTION 1.1. Background Research The growing demand and interest in beverages with health benefits and special characteristics has led to development of new functional beverages in order to increase a healthier lifestyle in society. Nowadays, stress, fatigue, and loss of vitality are just a few examples that happen in society. There are many important role of functional beverages in health promotion and disease prevention, for example can boosting the immunity system, relieving stress, improving intestinal health, and many more. Functional beverage has beneficial effects or more specific body functions and nutrition (Shahidi, 2004) The functional beverage in market covers many categories, from sports/energy drinks to herbal teas and fruit and vegetable juices, which targeted for all ages category because of that many health benefits. Beverages can be an excellent medium for the supplementation of nutritional components such as soluble fiber or herbal extract. The new formulations of beverages are rapidly changing. Nowadays, the market shelves are full with many different and new formulation of beverages such as soda pop, juices and dairy beverages, but some products taken as functional beverages such as coffees, teas, sports drinks, , probiotic drinks, smoothies and vegetable juices (Din et al., 2011). Probiotic drinks are one of functional beverage with added probiotic bacteria. The beneficial effects of food and beverage with added live microbes (probiotics) on human health are being increasingly promoted by health professionals (Schmidl & Labuza, 2000) Probiotic products that available on the market today are usually in the form of fermented milk and yoghurt. However, with an increase in vegan lifestyle throughout developed countries, there is also a demand for vegetarian probiotic products. -

L (+) Lactic Acid Fermentation and Its Product Polymerization
Electronic Journal of Biotechnology ISSN: 0717-3458 Vol.7 No.2, Issue of August 15, 2004 © 2004 by Pontificia Universidad Católica de Valparaíso -- Chile Received May 09, 2004 / Accepted June 10, 2004 REVIEW ARTICLE L (+) lactic acid fermentation and its product polymerization Niju Narayanan Department of Biochemical Engineering and Biotechnology Indian Institute of Technology Delhi Hauz Khas, New Delhi – 110016, India Tel: 91-11-26591001 Fax: 91-11-26582282 E-mail: [email protected] Pradip K. Roychoudhury Department of Biochemical Engineering and Biotechnology Indian Institute of Technology Delhi Hauz Khas, New Delhi – 110016, India Tel: 91-11-26591011 Fax: 91-11-26582282 E-mail: [email protected] Aradhana Srivastava* Department of Biochemical Engineering and Biotechnology Indian Institute of Technology Delhi Hauz Khas, New Delhi – 110016, India Tel: 91-11-26596192 Fax: 91-11-26582282 E-mail: [email protected] Keywords: bioprocesses, fermentation, L (+) lactic acid, Lactobacillus sp., polymerization. Lactic acid has been first introduced to us as early as Massachusetts, USA in 1881. Lactic acid can be 1780 as a sour component of milk. Ever since we have manufactured by (a) Chemical synthesis or (b) found its applications in food, pharmaceutical, cosmetic Carbohydrate fermentation. industries etc. Now there are emerging uses as a potential feedstock for the biodegradable polymer CHEMICAL SYNTHESIS industry. The microorganisms being used for lactic acid fermentation, the raw materials reported, the various The commercial process for chemical synthesis is based on novel fermentation processes and its processing methods lactonitrile. Hydrogen cyanide is added to acetaldehyde in have been reviewed. The properties and applications of the presence of a base to produce lactonitrile. -

Applications
Applications Lactic acid fermentation is used in many areas of the world to produce foods that cannot be produced through other methods[The most commercially important genus of lactic acid- fermenting bacteria is Lactobacillus, though other bacteria and even yeast are sometimes used[]Two of the most common applications of lactic acid fermentation are in the production of yogurt and sauerkraut. Pickle A product prepared by lactic acid bacteria (LAB) fermentation of sugars present in the pieces of fruits and vegetables. Traditionally, non-iodized salt is introduced to the vegetables through a brine, which inhibits spoilage but allows the growth of lactobacillus. The prepared product is rich in lactic acid, and only the beneficial bacteria that can tolerate lactic acid pH survive. It not only assures good quality of nutrients, but it is also a good source of probiotics. Fermented fish In some Asian cuisines, fish is traditionally fermented with rice to produce lactic acid that preserves the fish. Examples of these dishes include burong isda of the Philippines; narezushi of Japan; and pla ra of Thailand. The same process is also used for shrimp in the Philippines in the dish known as balao-balao Kimchi Kimchi also uses lactic acid fermentation Sauerkraut Lactic acid fermentation is also used in the production of sauerkraut. The main type of bacteria used in the production of sauerkraut is of the genus Leuconostoc As in yogurt, when the acidity rises due to lactic acid-fermenting organisms, many other pathogenic microorganisms are killed. The bacteria produce lactic acid, as well as simple alcohols and other hydrocarbons. -

FERMENTATION Differs from Respiration in Redox Considerations and Mechanism of ATP Synthesis No Exogenous Electron Accep
FERMENTATION Differs from respiration in redox considerations and mechanism of ATP synthesis No exogenous electron acceptors In anoxic habitats, if electron acceptors like sulfate, nitrate, ferric iron are in inadequate quantities, then organic compounds are catabolized by fermentation ATP is synthesized by substrate level phosphorylation Energy-rich compounds involved in substrate-level phosphorylation Compound Free energy of hydrolysis Acetyl-CoA -35.7 Propionyl-CoA -35.6 Butyryl-CoA -35.6 Succinyl-CoA -35.1 Acetyl phosphate -44.8 1,3-bisphosphoglycerate -51.9 Phosphoenolpyruvate -51.6 Adenosine -88.0 phosphosulfate Energy of hydrolysis of ATP (ATP ADP + Pi = -31.8 kJ/mol) Anaerobic metabolism of glucose Pyruvate decarboxylase is a key enzyme of ethanol fermentation.in Zymomonas (bacteria) and yeast Ethanol is produced through different reactions in saccharolytic clostridia, heterofermentative lactic acid bacteria and enteric bacteria. These bacteria oxidize pyruvate to acetyl-CoA before reducing it to ethanol. They do not possess pyruvate decarboxylase Fermentative diversity of bacteria Lactic acid fermentation by Lactic acid bacteria (LAB) Gram positive rods and cocci Grow anaerobically, but are aerotolerant Produce lactate as a major fermentation product Some LAB produce only lactate from sugars while others produce acetate and ethanol in addition to lactate. The former are referred to as homofermentative and the latter heterofermentative LAB. Homofermentative LAB ferment sugars through the EMP pathway Heterofermentative LAB ferment sugars through the phosphoketolase pathway Homolactate fermentation Homofermentative LAB include most species of Lactobacillus, Sporolactobacillus, Pediococcus, Enterococcus and Lactococcus. They use hexoses through the EMP pathway to generate ATP. Lactate dehydrogenase reoxidizes the NADH reduced during the EMP pathway using pyruvate as the electron acceptor As fermentation proceeds, lactate accumulates lowering the intracellular pH. -
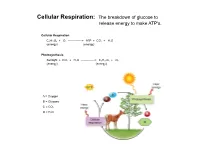
Cellular Respiration: the Breakdown of Glucose to Release Energy to Make ATP's
Cellular Respiration: The breakdown of glucose to release energy to make ATP's. Cellular Respiration C6H12O6 + O2 --------------> ATP + CO2 + H2O (energy) (energy) Photosynthesis Sunlight + CO2 + H2O --------------> C6H12O6 + O2 (energy) (energy) A = Oxygen B = Glucose C = CO2 D = H2O Cellular Respiration Overview Aerobic Respiration: The complete breakdown of glucose with oxygen resulting in lots of ATP's, CO2, H20. Phase 1: Glycolysis Reactions Reactions take place in the cytoplasm of cells. Reactants Products Glucose 4 ATP's 2 ATP's 2 NADH's 2 Pyruvic Acid molecules Phase 2: The Kreb's Cycle Reactions. Gets rid of Pyruvic Acid. Reactions take place in the fluid filled Matrix in a mitochondria. Reactant Products Total for 2 Pyruvic Acid Molecules Pyruvic Acid 4 NADH's 8 NADH's 1 FADH 2 FADH's 1 ATP 2 ATP'S CO2 Phase 3: The Electron Transport System (ETS) Reactions. Gets rid of NADH & FADH Reactions take place in the inner membrane Cristae of a mitochondria. Reactants Products NADH H2O FADH ATP's **** 2 ATP/FADH and 3 ATP/NADH O2 Aerobic Respiration totals for 1 Glucose molecule: From Glycolysis and Krebs 10 NADH's ----ETS------> 30 ATP's 2 FADH's ----ETS------> 4 ATP's ATP's from Glycolysis and Kreb's ---------> 6 ATP's Total ATP production for one molecule of glucose: 40 ATP (gross) Activiation energy needed for Glycolysis: -2 ATP Grand Total/Glucose: 38 ATP *Overall yield is probably around 36 ATP due to the active transport of Glycolysis' NADH into the mitochondria. Anaerobic Respiration: The incomplete breakdown of glucose WITHOUT OXYGEN resulting in a few ATP's and other high energy molecules. -

Notes: 9.3 - Fermentation
NOTES: 9.3 - FERMENTATION Vocabulary: -Fermentation -Glycolysis -Aerobic -Lactic Acid Fermentation -Anaerobic -Alcoholic Fermentation What happens if your cells aren’t getting enough oxygen? • Krebs Cycle & E.T.C. CANNOT proceed . • Instead, occurs -Pyruvic Acid builds up in & is converted to … feel the burn! -In other organisms, the pyruvic acid builds up, ferments, and becomes Fermentation: release of energy from food molecules in absence of oxygen ( ) • Enables cells to carry out energy production in the • result of glycolysis + fermentation yields 2 ATP molecules per 1 molecule of glucose (not nearly as much energy!!!!) 2 Fermentation Pathways 1) Alcoholic Fermentation 2) Lactic Acid Fermentation 1) Alcoholic Fermentation: • occurs in & some other microorganisms • pyruvic acid is broken down into a 2-Carbon plus • important to and ! -When yeast in dough runs out of oxygen it ferments, giving off bubbles of gas—which forms air space in bread -Alcohol produced in the dough evaporates when bread is baked *(when the level of alcohol reaches , yeast cells die) 2) Lactic Acid Fermentation: • pyruvic acid from glycolysis is • lactic acid is produced in muscles during rapid exercise (body can't supply enough O2 to the tissues) • the build-up of lactic acid causes a Energy and Exercise: Quick Energy: • Muscle cells only contain enough ATP for a few seconds of energy, or a “burst” ofenergy -e.g.: • When this is gone, cells resort to lactic acid fermentation which can provide enough ATP for about Energy and Exercise: Long Term Energy: • If a race or energy need is longer, your body must go through cellular respiration to get enough ATP to continue -even well-conditioned athletes have to pace themselves -your body stores energy in & in the form of .