The Rising Field of Bioinformatics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cognitive Science 1
Cognitive Science 1 • Linguistics, Minor (https://e-catalogue.jhu.edu/arts-sciences/full- COGNITIVE SCIENCE time-residential-programs/degree-programs/cognitive-science/ linguistics-minor/) http://www.cogsci.jhu.edu For current course information and registration go to https://sis.jhu.edu/ Cognitive science is the study of the human mind and brain, focusing on classes/ how the mind represents and manipulates knowledge and how mental representations and processes are realized in the brain. Conceiving of the mind as an abstract computing device instantiated in the brain, Courses cognitive scientists endeavor to understand the mental computations AS.050.102. Language and Mind. 3 Credits. underlying cognitive functioning and how these computations are Introductory course dealing with theory, methods, and current research implemented by neural tissue. Cognitive science has emerged at the topics in the study of language as a component of the mind. What it is interface of several disciplines. Central among these are cognitive to "know" a language: components of linguistic knowledge (phonetics, psychology, linguistics, and portions of computer science and artificial phonology, morphology, syntax, semantics) and the course of language intelligence; other important components derive from work in the acquisition. How linguistic knowledge is put to use: language and the neurosciences, philosophy, and anthropology. This diverse ancestry brain and linguistic processing in various domains. has brought into cognitive science several different perspectives and Area: Natural Sciences, Social and Behavioral Sciences methodologies. Cognitive scientists endeavor to unite such varieties AS.050.105. Introduction to Cognitive Neuropsychology. 3 Credits. of perspectives around the central goal of characterizing the structure When the brain is damaged or fails to develop normally, even the most of human intellectual functioning. -

Reliability in Cognitive Neuroscience: a Meta-Meta-Analysis Books Written by William R
Reliability in Cognitive Neuroscience: A Meta-Meta-Analysis Books Written by William R. Uttal Real Time Computers: Techniques and Applications in the Psychological Sciences Generative Computer Assisted Instruction (with Miriam Rogers, Ramelle Hieronymus, and Timothy Pasich) Sensory Coding: Selected Readings (Editor) The Psychobiology of Sensory Coding Cellular Neurophysiology and Integration: An Interpretive Introduction. An Autocorrelation Theory of Form Detection The Psychobiology of Mind A Taxonomy of Visual Processes Visual Form Detection in 3-Dimensional Space Foundations of Psychobiology (with Daniel N. Robinson) The Detection of Nonplanar Surfaces in Visual Space The Perception of Dotted Forms On Seeing Forms The Swimmer: An Integrated Computational Model of a Perceptual-Motor System (with Gary Bradshaw, Sriram Dayanand, Robb Lovell, Thomas Shepherd, Ramakrishna Kakarala, Kurt Skifsted, and Greg Tupper) Toward A New Behaviorism: The Case against Perceptual Reductionism Computational Modeling of Vision: The Role of Combination (with Ramakrishna Kakarala, Sriram Dayanand, Thomas Shepherd, Jaggi Kalki, Charles Lunskis Jr., and Ning Liu) The War between Mentalism and Behaviorism: On the Accessibility of Mental Processes The New Phrenology: On the Localization of Cognitive Processes in the Brain A Behaviorist Looks at Form Recognition Psychomyths: Sources of Artifacts and Misrepresentations in Scientifi c Cognitive neuroscience Dualism: The Original Sin of Cognitivism Neural Theories of Mind: Why the Mind-Brain Problem May Never Be Solved Human Factors in the Courtroom: Mythology versus Science The Immeasurable Mind: The Real Science of Psychology Time, Space, and Number in Physics and Psychology Distributed Neural Systems: Beyond the New Phrenology Neuroscience in the Courtroom: What Every Lawyer Should Know about the Mind and the Brain Mind and Brain: A Critical Appraisal of Cognitive Neuroscience Reliability in Cognitive Neuroscience: A Meta-Meta-Analysis Reliability in Cognitive Neuroscience: A Meta-Meta-Analysis William R. -

The Place of Modeling in Cognitive Science
The place of modeling in cognitive science James L. McClelland Department of Psychology and Center for Mind, Brain, and Computation Stanford University James L. McClelland Department of Psychology Stanford University Stanford, CA 94305 650-736-4278 (v) / 650-725-5699 (f) [email protected] Running Head: Modeling in cognitive science Keywords: Modeling frameworks, computer simulation, connectionist models, Bayesian approaches, dynamical systems, symbolic models of cognition, hybrid models, cognitive architectures Abstract I consider the role of cognitive modeling in cognitive science. Modeling, and the computers that enable it, are central to the field, but the role of modeling is often misunderstood. Models are not intended to capture fully the processes they attempt to elucidate. Rather, they are explorations of ideas about the nature of cognitive processes. As explorations, simplification is essential – it is only through simplification that we can fully understand the implications of the ideas. This is not to say that simplification has no downsides; it does, and these are discussed. I then consider several contemporary frameworks for cognitive modeling, stressing the idea that each framework is useful in its own particular ways. Increases in computer power (by a factor of about 4 million) since 1958 have enabled new modeling paradigms to emerge, but these also depend on new ways of thinking. Will new paradigms emerge again with the next 1,000-fold increase? 1. Introduction With the inauguration of a new journal for cognitive science, thirty years after the first meeting of the Cognitive Science Society, it seems essential to consider the role of computational modeling in our discipline. -

Author: Edwin Hutchins Title: Cognitive Ecology Affiliation: Department of Cognitive Science, University of California San Dieg
Author: Edwin Hutchins Title: Cognitive Ecology Affiliation: Department of Cognitive Science, University of California San Diego Tel: 858 534-1134 Fax: 858 822-2476 email: [email protected] Running Head: Cognitive Ecology Abstract: Cognitive ecology is the study of cognitive phenomena in context. In particular, it points to the web of mutual dependence among the elements of a cognitive ecosystem. At least three fields were taking a deeply ecological approach to cognition thirty years ago: Gibson’s ecological psychology, Bateson’s ecology of mind, and Soviet cultural-historical activity theory. The ideas developed in those projects have now found a place in modern views of embodied, situated, distributed cognition. As cognitive theory continues to shift from units of analysis defined by inherent properties of the elements to units defined in terms of dynamic patterns of correlation across elements, the study of cognitive ecosystems will become an increasingly important part of cognitive science. Keywords: units of analysis for cognition, ecological psychology, ecology of mind, activity theory, embodied cognition, situated cognition, distributed cognition, brain-body-world systems, human culture. Cognitive Ecology Choosing units of analysis for cognition Cognitive ecology is the study of cognitive phenomena in context. Elements of cognitive ecology have been present in various corners, but not the core, of cognitive science since the birth of the field. It is now being rediscovered as cognitive science shifts from viewing cognition as a logical process to seeing it as a biological phenomenon. Everything is connected to everything else. Fortunately, not all connectivity is equally dense. The non- uniformity of connectivity makes science possible. -

Research on the Strategy of E-Commerce Teaching Reform Based on Brain Cognition
KURAM VE UYGULAMADA EĞİTİM BİLİMLERİ EDUCATIONAL SCIENCES: THEORY & PRACTICE Received: November 2, 2017 Revision received: April 12, 2018 Copyright © 2018 EDAM Accepted: April 23, 2018 www.estp.com.tr DOI 10.12738/estp.2018.5.063 ⬧ October 2018 ⬧ 18(5) ⬧ 1637-1646 Research Article Research on the Strategy of E-commerce Teaching Reform Based on Brain Cognition 1 2 Ronggang Zhang Xiaheng Zhang Northwest University of Political Northwest University of Political Science and Law Science and Law Abstract The rapid development of brain science and cognitive science and the demand of education and teaching reform urge the development of brain-based education movement. With e-commerce teaching reform strategy as the research target, and literature analysis method, case teaching method and questionnaire as research methods, this paper constructs the teaching design pattern based on brain cognition of the course of Introduction to E- Commerce on the basis of the detailed analysis of teaching principle based on brain, teaching theory related to brain and theories related to teaching design, taking the course of Introduction to E-Commerce in colleges and universities as an example. The investigation and analysis of the course implementation and teaching effect verify that the teaching design pattern of e-commerce course based on brain cognition can promote students' interest in learning and knowledge mastery, providing a new teaching mode to be referred for e-commerce teaching reform. Keywords Brain Science • E-commerce Teaching • Teaching Elements • Teaching Design _______________________________________________ 1 Correspondence to: Ronggang Zhang (PhD), Business School, Northwest University of Political Science and Law, Xi’an710122, China. Email: [email protected] 2 Business School, Northwest University of Political Science and Law, Xi’an710122, China. -

The Cognitive Revolution: a Historical Perspective
Review TRENDS in Cognitive Sciences Vol.7 No.3 March 2003 141 The cognitive revolution: a historical perspective George A. Miller Department of Psychology, Princeton University, 1-S-5 Green Hall, Princeton, NJ 08544, USA Cognitive science is a child of the 1950s, the product of the time I went to graduate school at Harvard in the early a time when psychology, anthropology and linguistics 1940s the transformation was complete. I was educated to were redefining themselves and computer science and study behavior and I learned to translate my ideas into the neuroscience as disciplines were coming into existence. new jargon of behaviorism. As I was most interested in Psychology could not participate in the cognitive speech and hearing, the translation sometimes became revolution until it had freed itself from behaviorism, tricky. But one’s reputation as a scientist could depend on thus restoring cognition to scientific respectability. By how well the trick was played. then, it was becoming clear in several disciplines that In 1951, I published Language and Communication [1], the solution to some of their problems depended cru- a book that grew out of four years of teaching a course at cially on solving problems traditionally allocated to Harvard entitled ‘The Psychology of Language’. In the other disciplines. Collaboration was called for: this is a preface, I wrote: ‘The bias is behavioristic – not fanatically personal account of how it came about. behavioristic, but certainly tainted by a preference. There does not seem to be a more scientific kind of bias, or, if there is, it turns out to be behaviorism after all.’ As I read that Anybody can make history. -

Munin: a Peer-To-Peer Middleware for Ubiquitous Analytics and Visualization Spaces
IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS, VOL. XX, NO. Y, MONTH 2014 1 Munin: A Peer-to-Peer Middleware for Ubiquitous Analytics and Visualization Spaces Sriram Karthik Badam, Student Member, IEEE, Eli Fisher, and Niklas Elmqvist, Senior Member, IEEE Abstract—We present Munin, a software framework for building ubiquitous analytics environments consisting of multiple input and output surfaces, such as tabletop displays, wall-mounted displays, and mobile devices. Munin utilizes a service-based model where each device provides one or more dynamically loaded services for input, display, or computation. Using a peer-to-peer model for communication, it leverages IP multicast to replicate the shared state among the peers. Input is handled through a shared event channel that lets input and output devices be fully decoupled. It also provides a data-driven scene graph to delegate rendering to peers, thus creating a robust, fault-tolerant, decentralized system. In this paper, we describe Munin’s general design and architecture, provide several examples of how we are using the framework for ubiquitous analytics and visualization, and present a case study on building a Munin assembly for multidimensional visualization. We also present performance results and anecdotal user feedback for the framework that suggests that combining a service-oriented, data-driven model with middleware support for data sharing and event handling eases the design and execution of high performance distributed visualizations. Index Terms—Ubiquitous analytics, high-resolution displays, multi-display environments, distributed visualization, framework. F 1 INTRODUCTION peers can subscribe and publish, as well as a shared ISUALIZATION has long relied on computing devices event channel where peers can produce and consume V equipped with mice, keyboards and monitors for real-time events for input and coordination. -

Conceptual Framework for Quantum Affective Computing and Its Use in Fusion of Multi-Robot Emotions
electronics Article Conceptual Framework for Quantum Affective Computing and Its Use in Fusion of Multi-Robot Emotions Fei Yan 1 , Abdullah M. Iliyasu 2,3,∗ and Kaoru Hirota 3,4 1 School of Computer Science and Technology, Changchun University of Science and Technology, Changchun 130022, China; [email protected] 2 College of Engineering, Prince Sattam Bin Abdulaziz University, Al-Kharj 11942, Saudi Arabia 3 School of Computing, Tokyo Institute of Technology, Yokohama 226-8502, Japan; [email protected] 4 School of Automation, Beijing Institute of Technology, Beijing 100081, China * Correspondence: [email protected] Abstract: This study presents a modest attempt to interpret, formulate, and manipulate the emotion of robots within the precepts of quantum mechanics. Our proposed framework encodes emotion information as a superposition state, whilst unitary operators are used to manipulate the transition of emotion states which are subsequently recovered via appropriate quantum measurement operations. The framework described provides essential steps towards exploiting the potency of quantum mechanics in a quantum affective computing paradigm. Further, the emotions of multi-robots in a specified communication scenario are fused using quantum entanglement, thereby reducing the number of qubits required to capture the emotion states of all the robots in the environment, and therefore fewer quantum gates are needed to transform the emotion of all or part of the robots from one state to another. In addition to the mathematical rigours expected of the proposed framework, we present a few simulation-based demonstrations to illustrate its feasibility and effectiveness. This exposition is an important step in the transition of formulations of emotional intelligence to the quantum era. -

Cognitive Science for Computational Linguists
COGNITIVE SCIENCE FOR COMPUTATIONAL LINGUISTS ........................................................................................................................................... Overview Cognitive science is the scientific study of the human mind. The field is highly interdisciplinary, combining ideas and methods from psychology, computer science, linguistics, philosophy, and neuroscience. Broadly speaking, the goal is to characterize the nature of human knowledge ‐‐ its forms and content ‐‐ and how that knowledge is used, processed, and acquired. Active areas of cognitive research include language, memory, visual perception and cognition, thinking and reasoning, social cognition, decision making, and cognitive development. Important insights into issues of these kinds have been achieved in recent years as a result of the various Cognitive Science disciplines converging in their theoretical and methodological approaches. It is this convergence, in fact, that warrants grouping the disciplines together under the single term “Cognitive Science.” Even greater progress can be expected in the future as a consequence of increasing cooperation among the disciplines. Course participants will learn these topics through lectures and hands‐on experiments. Also case studies and assignments will be shared to stimulate research motivation of participants. Modules Course Duration: December 08‐14, 2016 This course will cover following topics: 1 : Introduction to Cognitive Science and Neuroscience 2 : Perception and attention Visual and auditory perception attention, Eye‐tracking. Applications of eye‐tracking in HCI. 3 :Conceptual representations in the mind and the brain. Concepts: Prototype theory, concept acquisition. The neural representation ofconcepts. 4 : Sentence processing Ambiguity, incrementality and disambiguation in lexical processing, syntax processing, and anaphoric processing, ERPs. 5 : Emotions in the mind and the brain Theories of emotions. The neural representation of emotions. Applications in designing artificial agents. -
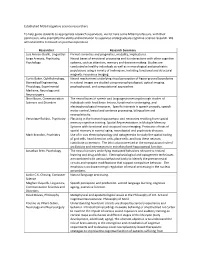
Established Cognitive Science Researchers
Established McGill cognitive science researchers To help guide students to appropriate research supervisors, we list here some McGill professors, with their permission, who exemplify the ability and inclination to supervise undergraduate cognitive science research. We will extend this list based on positive experience. Researcher Research Summary Luis Alonso-Ovalle, Linguistics Formal semantics and pragmatics, modality, implicatures. Jorge Armony, Psychiatry, Neural bases of emotional processing and its interactions with other cognitive Psychology systems, such as attention, memory and decision-making. Studies are conducted in healthy individuals as well as in neurological and psychiatric populations using a variety of techniques, including functional and structural magnetic resonance imaging. Curtis Baker, Ophthalmology, Neural mechanisms underlying visual perception of figure-ground boundaries Biomedical Engineering, in natural images are studied using neurophysiological, optical imaging, Physiology, Experimental psychophysical, and computational approaches Medicine, Neurology and Neurosurgery Shari Baum, Communication The neural bases of speech and language processing through studies of Sciences and Disorders individuals with focal brain lesions, functional neuroimaging, and electrophysiological measures. Specific interests in speech prosody, speech motor control, lexical and sentence processing, bilingualism and neuroplasticity. Veronique Bohbot, Psychiatry Plasticity in the human hippocampus and neocortex resulting from spatial memory cognitive training. Spatial Representations in Multiple Memory Systems with functional and structural neuroimaging. Protective effects of spatial memory in normal aging, neurological and psychiatric diseases. Mark Brandon, Psychiatry Use of in vivo electrophysiology and optogenetics to study the spatial coding of grid cells, head direction cells, place cells; and how these neurons contribute to memory. The lab is also interested in the computational role of oscillations and interneurons in entorhinal and hippocampal function. -

Reliability of Fmri Time Series: Similarity of Neural Processing During Movie Viewing
bioRxiv preprint doi: https://doi.org/10.1101/158188; this version posted July 3, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Reliability of fMRI time series: Similarity of neural processing during movie viewing Ralf Schm¨alzle1,2*, Martin A. Imhof2, Clare Grall1, Tobias Flaisch2, Harald T. Schupp2 1 Department of Communication, Michigan State University, USA 2 Department of Psychology, University of Konstanz, Germany Abstract Despite its widespread use in neuroscience, the reliability of fMRI remains insufficiently understood. One powerful way to tap into aspects of fMRI reliability is via the inter-subject correlation (ISC) approach, which exposes different viewers to the same time-locked naturalistic stimulus and assesses the similarity of neural time series. Here we examined the correlations of fMRI time series from 24 participants who watched the same movie clips across three repetitions. This enabled us to examine inter-subject correlations, intra-subject correlations, and correlations between aggregated time series, which we link to the notions of inter-rater reliability, stability, and consistency. In primary visual cortex we found average pairwise inter-subject correlations of about r = 0.3, and intra-subject correlations of similar magnitude. Aggregation across subjects increased inter-subject (inter-group) correlations to r = 0.87, and additional intra-subject averaging before cross-subject aggregation yielded correlations of r = 0.93. Computing the same analyses for parietal (visuospatial network) and cingulate cortices (saliency network) revealed a gradient of decreasing ISC from primary visual to higher visual to post-perceptual regions. -

Computation Vs. Information Processing: How They Are Different and Why It Matters
Studies in History and Philosophy of Science 41 (2010) 237–246 Contents lists available at ScienceDirect Studies in History and Philosophy of Science journal homepage: www.elsevier.com/locate/shpsa Computation vs. information processing: why their difference matters to cognitive science Gualtiero Piccinini a, Andrea Scarantino b a Department of Philosophy, University of Missouri St. Louis, 599 Lucas Hall (MC 73), One University Boulevard, St. Louis, MO 63121-4499, USA b Department of Philosophy, Neuroscience Institute, Georgia State University, PO Box 4089, Atlanta, GA 30302-4089, USA article info abstract Keywords: Since the cognitive revolution, it has become commonplace that cognition involves both computation and Computation information processing. Is this one claim or two? Is computation the same as information processing? The Information processing two terms are often used interchangeably, but this usage masks important differences. In this paper, we Computationalism distinguish information processing from computation and examine some of their mutual relations, shed- Computational theory of mind ding light on the role each can play in a theory of cognition. We recommend that theorists of cognition be Cognitivism explicit and careful in choosing notions of computation and information and connecting them together. Ó 2010 Elsevier Ltd. All rights reserved. When citing this paper, please use the full journal title Studies in History and Philosophy of Science 1. Introduction cessing, and the manipulation of representations does more