Susceptibility Tests the Mics Were Determined by an Agar Dilution Method Using Modified Gamagar (Nissui Seiyaku VOL.45 NO.4 the JOURNAL of ANTIBIOTICS 543
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-
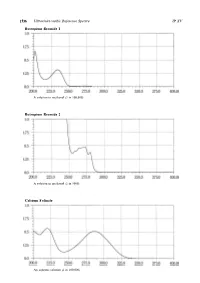
Ultraviolet-Visible Reference Spectra C
1536 Ultraviolet-visible Reference Spectra JP XV Butropium Bromide 1 A solution in methanol (1 in 100,000) Butropium Bromide 2 A solution in methanol (1 in 5000) Calcium Folinate An aqueous solution (1 in 100,000) JP XV Ultraviolet-visible Reference Spectra 1537 Camostat Mesilate An aqueous solution (1 in 100,000) Carbamazepine The sample solution obtained in the Assay Carbazochrome Sodium Sulfonate Hydrate An aqueous solution (1 in 100,000) 1538 Ultraviolet-visible Reference Spectra JP XV Carbidopa Hydrate A solution prepared as follows: Dissolve 0.01 g in 250 mL of a solution of hydrochloric acid in methanol (9 in 1000) Carmofur A solution in a mixture of methanol and phosphoric acid-acetic acid-boric acid buffer solution, pH 2.0 (9:1) (1 in 100,000) Carteolol Hydrochloride An aqueous solution (1 in 100,000) JP XV Ultraviolet-visible Reference Spectra 1539 Carumonam Sodium An aqueous solution (3 in 100,000) Cefaclor An aqueous solution (1 in 50,000) Cefadroxil An aqueous solution (1 in 50,000) 1540 Ultraviolet-visible Reference Spectra JP XV Cefalexin An aqueous solution (3 in 100,000) Cefalotin Sodium An aqueous solution (1 in 50,000) Cefapirin Sodium An aqueous solution (3 in 200,000) JP XV Ultraviolet-visible Reference Spectra 1541 Cefatrizine Propylene Glycolate An aqueous solution (1 in 50,000) Cefazolin Sodium An aqueous solution (1 in 50,000) Cefbuperazone Sodium An aqueous solution (1 in 50,000) 1542 Ultraviolet-visible Reference Spectra JP XV Cefditoren Pivoxil A solution in methanol (1 in 50,000) Cefixime A solution in 0.1 -

00246-2019.Supptables
1 Table S1. List of ICD-10 codes identifying comorbidities during hospitalization for COPD exacerbation COPD J41, J42, J43, J440, J441, J449 COPD exacerbation J10, J11, J13, J14, J15, J16, J18, J20, J21, J22, J46, J170, J171, J178, J440, J441, J851, J12, A481, B012, B052, B250 Bacterial pneumonia J100, J110, J12, J13, J14, J15, J16, J170, J178, J18, J851, A481 Lung cancer C34 Other malignancy C00-26, C30-33, C37-41, C43-58, C60-76 Diabetes/abnormal glucose tolerance E10-14, R703 Bone fracture/osteoporosis M80-84 Interstitial pneumonia B221, J701, J704, J841, J848, J849, J990, J991, M321, M330, M331, M332, M351 Bronchial asthma J45, J46 Bronchiectasis J40, J41, J42, J47 Pulmonary thromboembolism I26 Mycobacterium infection A150-154, A156-159, A161-162, A165, A168-169, A19, A310, A319 Mycotic infection A420, A43, B37, B380-382, B390-392, B400-402, B410, B420, B440, B441, B449, B460, J172 Cor pulmonare I27 Congestive heart failure E059, I46, I50, I099, I110 Ischemic heart disease I20-25 Tachycardia I47-49, R000, T818 Autoimmune disease M05, M06, M08, M30-35 Stroke I60-64 Liver dysfunction B89, B181, B182, B659, B661, K702, K703, K72, K74, K761, K762, K763, K766, K767 Renal failure E102, E112, E142, I120, N17-19 GERD K21 Constipation or ileus K56, K590 Prostate hypertrophy N40 Abbreviations: COPD, chronic obstructive pulmonary disease; GERD, gastro-esophageal reflux disease 2 Table S2. List of baseline characteristics, comorbidities, and treatments before and during hospitalization for COPD exacerbation Baseline characteristics Sex, fiscal -

Breeding Laboratories (Hino, Japan)
VOL. XLII NO. 6 THE JOURNAL OF ANTIBIOTICS 989 STIMULATORY EFFECT OF CEFODIZIME ON MACROPHAGE- MEDIATED PHAGOCYTOSIS Kazunori Oishi, Keizo Matsumoto, Masashi Yamamoto, Toshihiro Morito and Toshiaki Yoshida Department of Internal Medicine, Institute of Tropical Medicine, Nagasaki University, 12-4 Sakamoto-machi, Nagasaki 852, Japan (Received for publication January 27, 1989) Weevaluated the ingestion of anti-sheep erythrocyte (anti-E) IgG- and IgM-coated sheep erythiocytes by murineperitoneal macrophagesexposed to cefodizime, a newsemisynthetic cephalosporin, and other antibiotics. Cefodizime enhanced the ingestion of anti-E IgG- coated erythrocytes by peritoneal macrophages from CD-I and BALB/cmice in a dose- dependent manner, but had no effect on uncoated or IgM-coated erythrocytes. Similar enhancement was observed only in the case of cefpimizole (AC-1370), among the other anti- biotics examined. These results suggest that the favorable in vivo activity of cefodizime and cefpimizole may result from their phagocytosis-enhancing as well as antimicrobial properties. The new semisynthetic cephalosporin, cefodizime, is characterized by a cephem ring which con- tains a ^w-methoxyimino-aminothiazolyl group at the 7-position and a thiazolylthio-methyl group at the 3-position. The latter substitution is thought to provide metabolic stability and a prolonged half-life in humanserum0. The efficacy of cefodizime in experimental murine infections is apparently superior to that predicted from in vitro activity2>3). Wepostulated that the enhanced in vivo activity of cefodizime may be due to drug-induced immunostimulation. This speculation was in part based upon the previous demonstration that a variety of agents, including lysophosphatidylcholine4) and fibronectin5), enhance receptor-dependent phagocytosis. Moreover, it was previously reported that cefpimizole (AC-1370), another semisynthetic cephalosporin, potentiated phagocyte function of macrophages and neutrophils6). -

Consideration of Antibacterial Medicines As Part Of
Consideration of antibacterial medicines as part of the revisions to 2019 WHO Model List of Essential Medicines for adults (EML) and Model List of Essential Medicines for children (EMLc) Section 6.2 Antibacterials including Access, Watch and Reserve Lists of antibiotics This summary has been prepared by the Health Technologies and Pharmaceuticals (HTP) programme at the WHO Regional Office for Europe. It is intended to communicate changes to the 2019 WHO Model List of Essential Medicines for adults (EML) and Model List of Essential Medicines for children (EMLc) to national counterparts involved in the evidence-based selection of medicines for inclusion in national essential medicines lists (NEMLs), lists of medicines for inclusion in reimbursement programs, and medicine formularies for use in primary, secondary and tertiary care. This document does not replace the full report of the WHO Expert Committee on Selection and Use of Essential Medicines (see The selection and use of essential medicines: report of the WHO Expert Committee on Selection and Use of Essential Medicines, 2019 (including the 21st WHO Model List of Essential Medicines and the 7th WHO Model List of Essential Medicines for Children). Geneva: World Health Organization; 2019 (WHO Technical Report Series, No. 1021). Licence: CC BY-NC-SA 3.0 IGO: https://apps.who.int/iris/bitstream/handle/10665/330668/9789241210300-eng.pdf?ua=1) and Corrigenda (March 2020) – TRS1021 (https://www.who.int/medicines/publications/essentialmedicines/TRS1021_corrigenda_March2020. pdf?ua=1). Executive summary of the report: https://apps.who.int/iris/bitstream/handle/10665/325773/WHO- MVP-EMP-IAU-2019.05-eng.pdf?ua=1. -

Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DIX to the HTSUS—Continued
20558 Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DEPARMENT OF THE TREASURY Services, U.S. Customs Service, 1301 TABLE 1.ÐPHARMACEUTICAL APPEN- Constitution Avenue NW, Washington, DIX TO THE HTSUSÐContinued Customs Service D.C. 20229 at (202) 927±1060. CAS No. Pharmaceutical [T.D. 95±33] Dated: April 14, 1995. 52±78±8 ..................... NORETHANDROLONE. A. W. Tennant, 52±86±8 ..................... HALOPERIDOL. Pharmaceutical Tables 1 and 3 of the Director, Office of Laboratories and Scientific 52±88±0 ..................... ATROPINE METHONITRATE. HTSUS 52±90±4 ..................... CYSTEINE. Services. 53±03±2 ..................... PREDNISONE. 53±06±5 ..................... CORTISONE. AGENCY: Customs Service, Department TABLE 1.ÐPHARMACEUTICAL 53±10±1 ..................... HYDROXYDIONE SODIUM SUCCI- of the Treasury. NATE. APPENDIX TO THE HTSUS 53±16±7 ..................... ESTRONE. ACTION: Listing of the products found in 53±18±9 ..................... BIETASERPINE. Table 1 and Table 3 of the CAS No. Pharmaceutical 53±19±0 ..................... MITOTANE. 53±31±6 ..................... MEDIBAZINE. Pharmaceutical Appendix to the N/A ............................. ACTAGARDIN. 53±33±8 ..................... PARAMETHASONE. Harmonized Tariff Schedule of the N/A ............................. ARDACIN. 53±34±9 ..................... FLUPREDNISOLONE. N/A ............................. BICIROMAB. 53±39±4 ..................... OXANDROLONE. United States of America in Chemical N/A ............................. CELUCLORAL. 53±43±0 -

The Genus Ochrobactrum As Major Opportunistic Pathogens
microorganisms Review The Genus Ochrobactrum as Major Opportunistic Pathogens Michael P. Ryan 1,2 and J. Tony Pembroke 2,* 1 Department of Applied Sciences, Limerick Institute of Technology, Moylish V94 EC5T, Limerick, Ireland; [email protected] 2 Molecular Biochemistry Laboratory, Department of Chemical Sciences, School of Natural Sciences, Bernal Institute, University of Limerick, Limerick V94 T9PX2, Ireland * Correspondence: [email protected] Received: 22 October 2020; Accepted: 13 November 2020; Published: 16 November 2020 Abstract: Ochrobactrum species are non-enteric, Gram-negative organisms that are closely related to the genus Brucella. Since the designation of the genus in 1988, several distinct species have now been characterised and implicated as opportunistic pathogens in multiple outbreaks. Here, we examine the genus, its members, diagnostic tools used for identification, data from recent Ochrobactrum whole genome sequencing and the pathogenicity associated with reported Ochrobactrum infections. This review identified 128 instances of Ochrobactrum spp. infections that have been discussed in the literature. These findings indicate that infection review programs should consider investigation of possible Ochrobactrum spp. outbreaks if these bacteria are clinically isolated in more than one patient and that Ochrobactrum spp. are more important pathogens than previously thought. Keywords: Ochrobactrum; nosocomial infection; environmental bacteria 1. Introduction Gram-negative, non-fermenting bacteria are an emergent worry in medical situations and are becoming a growing cause of severe infections. Pathogens of this type are opportunistic and include many different bacterial species, such as Ralstonia spp., Pseudomonas aeruginosa, Sphingomonas paucimobilis and Brevundimonas spp. [1–5]. Gram-negative, non-fermenting bacteria can infect both patients undergoing treatments and individuals outside of a clinical setting with various underlying conditions or diseases. -

PHARMACEUTICAL APPENDIX to the HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2008) (Rev
Harmonized Tariff Schedule of the United States (2008) (Rev. 2) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2008) (Rev. 2) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 2 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. ABACAVIR 136470-78-5 ACIDUM GADOCOLETICUM 280776-87-6 ABAFUNGIN 129639-79-8 ACIDUM LIDADRONICUM 63132-38-7 ABAMECTIN 65195-55-3 ACIDUM SALCAPROZICUM 183990-46-7 ABANOQUIL 90402-40-7 ACIDUM SALCLOBUZICUM 387825-03-8 ABAPERIDONUM 183849-43-6 ACIFRAN 72420-38-3 ABARELIX 183552-38-7 ACIPIMOX 51037-30-0 ABATACEPTUM 332348-12-6 ACITAZANOLAST 114607-46-4 ABCIXIMAB 143653-53-6 ACITEMATE 101197-99-3 ABECARNIL 111841-85-1 ACITRETIN 55079-83-9 ABETIMUSUM 167362-48-3 ACIVICIN 42228-92-2 ABIRATERONE 154229-19-3 ACLANTATE 39633-62-0 ABITESARTAN 137882-98-5 ACLARUBICIN 57576-44-0 ABLUKAST 96566-25-5 ACLATONIUM NAPADISILATE 55077-30-0 ABRINEURINUM 178535-93-8 ACODAZOLE 79152-85-5 ABUNIDAZOLE 91017-58-2 ACOLBIFENUM 182167-02-8 ACADESINE 2627-69-2 ACONIAZIDE 13410-86-1 ACAMPROSATE -

(Penicillin Or Cephalosporin R3-Lactam Hydrolyzing Enzymes
VOL. XXXVII NO. 5 THE JOURNAL OF ANTIBIOTICS 577 INACTIVATION OF CEPHAMYCINS BY VARIOUS ,3-LACTAMASES FROM GRAM-NEGATIVE BACTERIA SHINZABUROU MINAMI, NOBUYUKI MATSUBARA,AKIRA YOTSUJI, HARUMI ARAKI, YASUO WATANABE, TAKASHI YASUDA, ISAMU SAIKAWA and SUSUMU MITSUHASHI' Research Laboratory, Toyama Chemical Co., Ltd., Toyama, Japan 'Department of Microbiology , School of Medicine, Gumma University, Maebashi, Japan (Received for publication November 10, 1983) The enzymic inactivation of cephamycins, i.e. cefoxitin, cefmetazole, cefotetan and cefbuperazone, was investigated by means of bioassay, high pressure liquid chromatography (HPLC) and spectrophotometric analysis using three types of cephalosporinase (CSase, RICHMONDtype la, lb and Ic) and one penicillinase (PCase, TEM type). These four cepha- mycins were not inactivated by Ic CSase and TEM type PCase or producers of these enzymes. However, the inactivation of cefmetazole and cefoxitin was noted when they were incubated in the cultures of CSase (la and 1b)-producers or incubated with a large amount of these purified enzymes although the inactivation of cefbuperazone was not noted. HPLC of culture fluid or enzyme solution which contained cefmetazole or cefoxitin and were incubated at 37°C showed that metabolites of cefmetazole or cefoxitin appeared as the drug disappeared. In addition, the appearance of metabolites corresponded to the loss of the drug's bioactivities and the absorption of iodine. UV and IR spectra of cefmetazole which were taken after incubation with the purified CSase -

In Vitro Activity Ofcefbuperazone Against Bacteroides Spp
ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, June 1985, p. 96-970 Vol. 27, No. 6 0066-4804/85/060968-03$02.00/0 Copyright © 1985, American Society for Microbiology In Vitro Activity of Cefbuperazone Against Bacteroides spp. M. BEATRIZ SOUZA DIAS, NILDA V. JACOBUS, SHERWOOD L. GORBACH, AND FRANCIS P. TALLY* Division ofInfectious Diseases, Department of Medicine, New England Medical Center Hospital, Boston, Massachusetts 02111 Received 9 November 1984/Accepted 14 March 1985 The activity of cefbuperazone was tested in vitro against 287 clinical isolates of Bacteroides spp. Cefbuperazone showed good activity against P. fragils, B. vulgatus, and other Bacteroides species, comparable to that of cefoxitin. It was relatively ineffective against B. distasonis and the B. thetaiotaomicron-ovatus group and was not active against cefoxitin-resistant Barteroides spp. Cefbuperazone (BMY-25182) is a new semisynthetic the B. thetaiotaomicron-ovatus group showed high rates of cephamycin antibiotic. It has a methylthiotetrazole ring at resistance to most ofthe drugs tested, especially to cefbuper- position 3 of the dihydrothiazine ring and a dioxopiperazine azone. Cefotaxime was the most active agent against B. ring as a side chain, a structure similar to those of cefoper- distasonis, and piperacillin was the most active against the azone and piperacillin. In addition, its ,B-lactam ring is B. thetaiotaomicron-ovatus group. Among the B. vulgatus protected against ,B-lactamase activity by the methoxy group strains tested, 83% were susceptible to cefbuperazone, cefox- at position 7 of the P-lactamn ring, as has been shown for itin, moxalactam, cefotaxime, and piperacillin, whereas only other cephamycin compounds (7). The present study com- 50% were susceptible to cefoperazone. -

(12) United States Patent (10) Patent No.: US 6,613,807 B2 Uhrich (45) Date of Patent: *Sep
USOO6613807B2 (12) United States Patent (10) Patent No.: US 6,613,807 B2 Uhrich (45) Date of Patent: *Sep. 2, 2003 (54) THERAPEUTIC POLYANHYDRIDE DE 288387 3/1991 ........... CO8G/67/04 COMPOUNDS FOR DRUG DELIVERY EP O246341 11/1987 ........... A61 L/27/00 EP O 246 341 * 11/1987 (75) Inventor: Kathryn E. Uhrich, Hoboken, NJ (US) NL 9000237 8/1991 .......... A61 K/31/60 WO WO 90/09779 9/1990 ............ A61K/7/16 WO 91/O9831 * 7/1991 (73) ASSignee: ST'State tly, New W. WO-91/09831 7/1991 ......... CO7C/69/035 ersey, New Brunswick, NJ (US) WO WO-97/39738 10/1997 ... A61 K/9/16 (*) Notice: Subject to any disclaimer, the term of this W Wo: to r : patent is extended or adjusted under 35 WO 99/12990 * 3/1999 U.S.C. 154(b) by 0 days. WO WO-99/29885 6/1999 ............. C12P/1/OO This patent is Subject to a terminal dis OTHER PUBLICATIONS claimer. Anastasiou, T.J. "Novel Polyanhydrides with Enhanced Thermal and Solubility Properties”, Macromolecules, 33 (21) Appl. No.: 09/917,231 (17), (2000).pp, 6217–6221. Anastasiou, T.J. "Novel, Degradable Polyanhydrides”, (22) Filed: Jul. 27, 2001 25th Annual Meeting Transactions of the Society for Bio (65) Prior Publication Data materials, Abstract,(1999).p, 79, Anastasiou, T.J. "Synthesis of Novel, Degradable Polyan US 2002/0098161 A1 Jul. 25, 2002 hydrides Containing Para-AminoSalicylic Acid as Drug Delivery Devices for Tuberculosis Treatment', Polymer Related U.S. Application Data Preprints, 41(2), (Aug. 2000),pp. 1366–1367. Attawia, M.A. "Biocompatibility Testing of Poly(anhy (63) Continuation-in-part of application No. -

Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use in European Acute Care Hospitals
TECHNICAL DOCUMENT Point prevalence survey of healthcare-associated infections and antimicrobial use in European acute care hospitals Protocol version 5.3 www.ecdc.europa.eu ECDC TECHNICAL DOCUMENT Point prevalence survey of healthcare- associated infections and antimicrobial use in European acute care hospitals Protocol version 5.3, ECDC PPS 2016–2017 Suggested citation: European Centre for Disease Prevention and Control. Point prevalence survey of healthcare- associated infections and antimicrobial use in European acute care hospitals – protocol version 5.3. Stockholm: ECDC; 2016. Stockholm, October 2016 ISBN 978-92-9193-993-0 doi 10.2900/374985 TQ-04-16-903-EN-N © European Centre for Disease Prevention and Control, 2016 Reproduction is authorised, provided the source is acknowledged. ii TECHNICAL DOCUMENT PPS of HAIs and antimicrobial use in European acute care hospitals – protocol version 5.3 Contents Abbreviations ............................................................................................................................................... vi Background and changes to the protocol .......................................................................................................... 1 Objectives ..................................................................................................................................................... 3 Inclusion/exclusion criteria .............................................................................................................................. 4 Hospitals ................................................................................................................................................. -

Statistical Analysis Plan / Data Specifications the Risk of Acute Liver Injury Associated with the Use of Antibiotics. a Replica
WP6 validation on methods involving an extended audience Statistical analysis plan / data specifications The risk of acute liver injury associated with the use of antibiotics. A replication study in the Utrecht Patient Oriented Database Version: July 11, 2013 Authors: Renate Udo, Marie L De Bruin Reviewers: Work package 6 members (David Irvine, Stephany Tcherny-Lessenot) 1 1. Context The study described in this protocol is performed within the framework of PROTECT (Pharmacoepidemiological Research on Outcomes of Therapeutics by a European ConsorTium). The overall objective of PROTECT is to strengthen the monitoring of the benefit-risk of medicines in Europe. Work package 6 “validation on methods involving an extended audience” aims to test the transferability/feasibility of methods developed in other WPs (in particular WP2 and WP5) in a range of data sources owned or managed by Consortium Partners or members of the Extended Audience. The specific aims of this study within WP6 are: to evaluate the external validity of the study protocol on the risk of acute liver injury associated with the use of antibiotics by replicating the study protocol in another database, to study the impact of case validation on the effect estimate for the association between antibiotic exposure and acute liver injury. Of the selected drug-adverse event pairs selected in PROTECT, this study will concentrate on the association between antibiotic use and acute liver injury. On this topic, two sub-studies are performed: a descriptive/outcome validation study and an association study. The descriptive/outcome validation study has been conducted within the Utrecht Patient Oriented Database (UPOD).