The Holliday Junction in an Inverted Repeat DNA Sequence: Sequence Effects on the Structure of Four-Way Junctions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An RNA Holliday Junction? Structural and Dynamic Considerations of the Bacteriophage T4 Gene 60 Interruption
J. Mol. Biol. (1990) 213, 199-201 COMMUNICATIONS An RNA Holliday Junction? Structural and Dynamic Considerations of the Bacteriophage T4 Gene 60 Interruption Donald H. Burke-Agiiero and John E. Hearst Department of Chemistry University of California, Laboratory of Chemical Biodynamics Lawrence Berkeley Laboratory, Berkeley, CA 94720, U.S.A. (Received 20 June 1988; accepted 20 November 1989) A novel interrupted gene motif has been reported in which a 50-nucleotide insertion into bacteriophage T4 gene 60 appears to be present in the message at the time of translation, yet it is not translated. We present here a dynamic model for how translation may be occurring in the neighborhood of the interruption. The model involves formation of an RNA structure with similarities to a Holliday junction, followed by migration of the branch point in a strand exchange between message and interruption. The advantage of this model over previous ones is that at no time is a new tRNA required to pair with a discontinuous template. Huang et al. (1988) have reported a novel gene tion; the order of events is not important to this interruption motif in gene 60 of bacteriophage T4, model. More extensive branch migration is hindered in which a 50-nucleotide intervening segment is by the lack of sequence homology between the neither removed nor translated. The sequence of the message and the dumbbell outside these five bases. interruption suggests that it can be folded to bring When tRNA G~y moves into the P site, the template the translated codons on either side of the inter- 3' to Gly46 will now be in position to receive the ruption into close proximity (Fig. -

Holliday Junction Resolvases
Downloaded from http://cshperspectives.cshlp.org/ on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press Holliday Junction Resolvases Haley D.M. Wyatt and Stephen C. West London Research Institute, Cancer Research UK, Clare Hall Laboratories, South Mimms, Herts EN6 3LD, United Kingdom Correspondence: [email protected] Four-way DNA intermediates, called Holliday junctions (HJs), can form during meiotic and mitotic recombination, and their removal is crucial for chromosome segregation. A group of ubiquitous and highly specialized structure-selective endonucleases catalyze the cleavage of HJs into two disconnected DNA duplexes in a reaction called HJ resolution. These enzymes, called HJ resolvases, have been identified in bacteria and their bacteriophages, archaea, and eukaryotes. In this review, we discuss fundamental aspects of the HJ structure and their interaction with junction-resolving enzymes. This is followed by a brief discussion of the eubacterial RuvABC enzymes, which provide the paradigm for HJ resolvases in other organisms. Finally, we review the biochemical and structural properties of some well-char- acterized resolvases from archaea, bacteriophage, and eukaryotes. omologous recombination (HR) is an es- homologous strand as a template for DNA syn- Hsential process that promotes genetic di- thesis. Recombination then proceeds in one of versity during meiosis (see Lam and Keeney several different ways, some of which involve 2014; Zickler and Kleckner 2014). However, in second-end capture, such that the other resect- somatic cells, HR plays a key role in conserv- ed 30 end anneals to the displaced strand of the ing genetic information by facilitating DNA re- D-loop (Szostak et al. -

Insights Into Holliday Junction Resolution and Chromosome Segregation in Yeast
September 17, 2012 SCIENCE SPOTLIGHT Insights into Holliday Junction Resolution and Chromosome Segregation in Yeast September 17, 2012 ME Arnegard Homologous DNA recombination is the highly conserved process by which similar or identical nucleotide sequences are exchanged between two DNA molecules. In all eukaryotes – from yeast to humans – this kind of genetic recombination serves two critical functions during meiosis, the class of cell division that leads to the formation of gametes such as sperm and eggs in the case of humans, or spores in yeast: Homologous recombination promotes the genetic diversity of gametes by allowing crossing-over between homologous chromosomes, and it is also critical to accurate chromosome segregation during meiosis. Anomalies in recombination often result in the complete loss of chromosomes during meiosis, or in chromosomal rearrangements such as deletions or translocations, which in turn can lead to birth defects or cancer. All current models for crossing-over involve two key intermediate structures: DNA double strand breaks (DSBs) and Holliday junctions (HJs). DSBs are the initiating events in crossing-over that are caused by Rec12 in fission yeast, whereas HJs are the crossed-strand structures by which DNA molecules from two homologous chromosomes become intertwined (see figure). At the Fred Hutchinson Cancer Research Center, the laboratory of Dr. Gerald Smith (Basic Sciences Division) has made many advances toward understanding a complex pathway composed of dozens of proteins that promote homologous recombination in fission yeast (Schizosaccharomyces pombe) by moving or pairing chromosomes, or forming or repairing DSBs. In a previous study, for example, Cromie et al. (2006) provided strong evidence that HJs in fission yeast are 'resolved' (i.e., precisely cut and reconnected into two independent helices) solely by the endonucleolytic activity of the partner proteins Mus81 and Eme1. -

Working with Molecular Genetics Chapter 8. Recombination of DNA CHAPTER 8 RECOMBINATION of DNA
Working with Molecular Genetics Chapter 8. Recombination of DNA CHAPTER 8 RECOMBINATION OF DNA The previous chapter on mutation and repair of DNA dealt mainly with small changes in DNA sequence, usually single base pairs, resulting from errors in replication or damage to DNA. The DNA sequence of a chromosome can change in large segments as well, by the processes of recombination and transposition. Recombination is the production of new DNA molecule(s) from two parental DNA molecules or different segments of the same DNA molecule; this will be the topic of this chapter. Transposition is a highly specialized form of recombination in which a segment of DNA moves from one location to another, either on the same chromosome or a different chromosome; this will be discussed in the next chapter. Types and examples of recombination At least four types of naturally occurring recombination have been identified in living organisms (Fig. 8.1). General or homologous recombination occurs between DNA molecules of very similar sequence, such as homologous chromosomes in diploid organisms. General recombination can occur throughout the genome of diploid organisms, using one or a small number of common enzymatic pathways. This chapter will be concerned almost entirely with general recombination. Illegitimate or nonhomologous recombination occurs in regions where no large- scale sequence similarity is apparent, e.g. translocations between different chromosomes or deletions that remove several genes along a chromosome. However, when the DNA sequence at the breakpoints for these events is analyzed, short regions of sequence similarity are found in some cases. For instance, recombination between two similar genes that are several million bp apart can lead to deletion of the intervening genes in somatic cells. -

Protein-Assisted Room-Temperature Assembly of Rigid, Immobile Holliday Junctions and Hierarchical DNA Nanostructures
molecules Article Protein-Assisted Room-Temperature Assembly of Rigid, Immobile Holliday Junctions and Hierarchical DNA Nanostructures 1,2, 3,4, 1, Saminathan Ramakrishnan y, Sivaraman Subramaniam y, Charlotte Kielar z, Guido Grundmeier 1 , A. Francis Stewart 3,4 and Adrian Keller 1,* 1 Technical and Macromolecular Chemistry, Paderborn University, Warburger Str. 100, 33098 Paderborn, Germany; [email protected] (S.R.); [email protected] (C.K.); [email protected] (G.G.) 2 Structural Biophysics Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA 3 Biotechnology Center, Department of Genomics, Technische Universität Dresden, Tatzberg 47-51, 01307 Dresden, Germany; [email protected] (S.S.); [email protected] (A.F.S.) 4 Cluster of Excellence Physics of Life, Technische Universität Dresden, 01062 Dresden, Germany * Correspondence: [email protected] These authors contributed equally to this work. y Present address: Institute of Resource Ecology, Helmholtz-Zentrum Dresden-Rossendorf, z Bautzner Landstraße 400, 01328 Dresden, Germany. Academic Editor: Ramon Eritja Received: 29 September 2020; Accepted: 30 October 2020; Published: 3 November 2020 Abstract: Immobile Holliday junctions represent not only the most fundamental building block of structural DNA nanotechnology but are also of tremendous importance for the in vitro investigation of genetic recombination and epigenetics. Here, we present a detailed study on the room-temperature assembly of immobile Holliday junctions with the help of the single-strand annealing protein Redβ. Individual DNA single strands are initially coated with protein monomers and subsequently hybridized to form a rigid blunt-ended four-arm junction. We investigate the efficiency of this approach for different DNA/protein ratios, as well as for different DNA sequence lengths. -

The Crystal Structures of DNA Holliday Junctions P Shing Ho* and Brandt F Eichman†
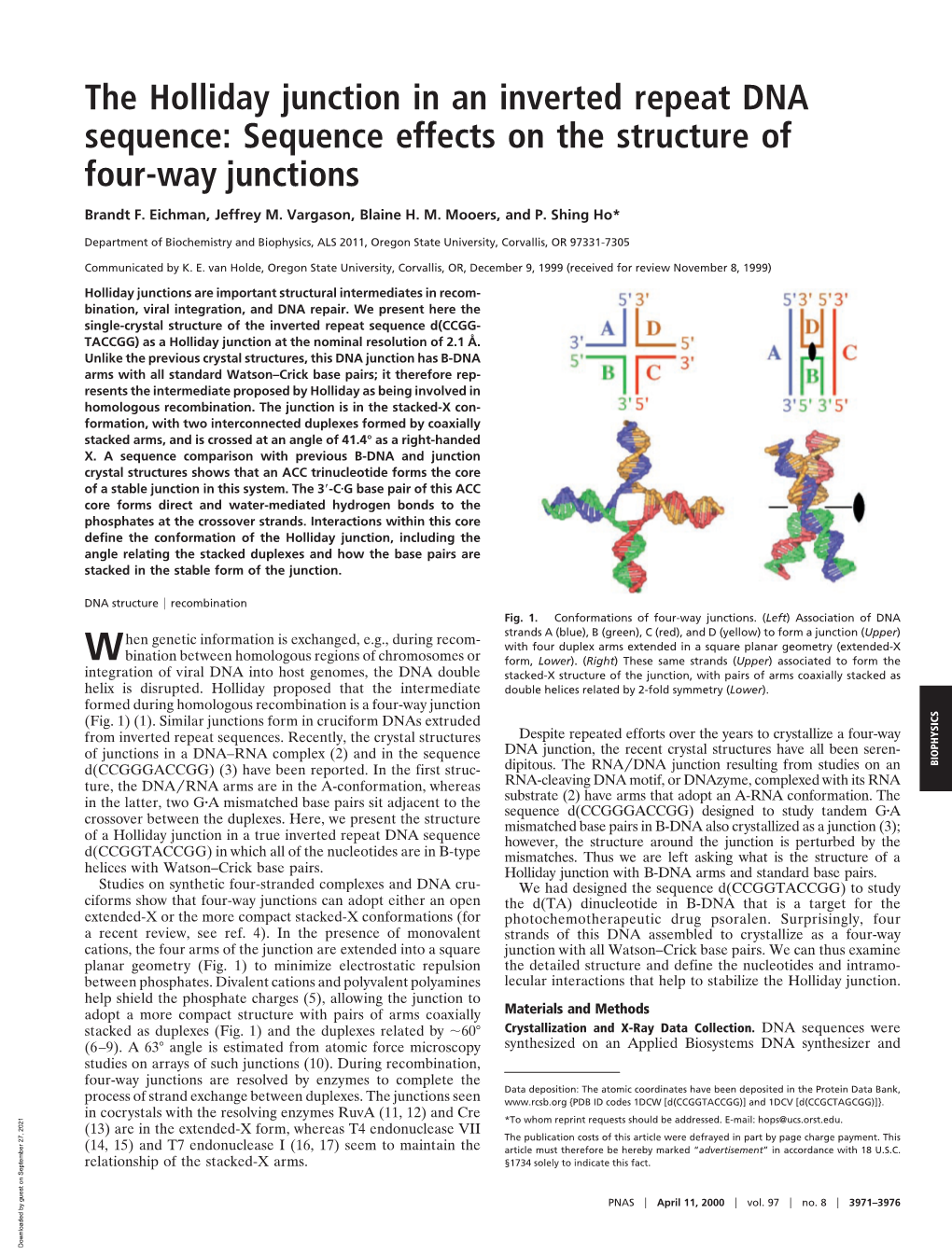
302 The crystal structures of DNA Holliday junctions P Shing Ho* and Brandt F Eichman† Nearly 40 years ago, Holliday proposed a four-stranded Molecular models of the four-way junction complex or junction as the central intermediate in the general It is now generally accepted that there are two structural mechanism of genetic recombination. During the past two forms of the Holliday junction (reviewed in [9•]). Low salt years, six single-crystal structures of such DNA junctions have conditions favor an open-X form, in which the four arms been determined by three different research groups. These are extended in a square planar geometry (Figure 1b), structures all essentially adopt the antiparallel stacked-X thereby minimizing the repulsion between the negatively conformation, but can be classified into three distinct charged phosphates concentrated at the junction. This is categories: RNA–DNA junctions; ACC trinucleotide junctions; also the form observed in complexes with the enzymes and drug-induced junctions. Together, these structures provide RuvA [10,11,12•] and Cre [13]. insight into how local and distant interactions help to define the detailed and general physical features of Holliday junctions In contrast, the presence of multivalent cations effective- at the atomic level. ly shields these negative charges, allowing the arms to coaxially pair and stack into the more compact stacked-X Addresses junction (Figure 1c). Before the recent crystal studies, *Department of Biochemistry and Biophysics, ALS 2011, Oregon the most detailed models of four-way junctions were State University, Corvallis, Oregon 97331, USA; derived from studies using gel electrophoresis, fluores- e-mail: [email protected] cence resonance energy transfer, NMR spectroscopy and †Department of Biological Chemistry and Molecular Pharmacology, • Harvard Medical School, Boston, Massachusetts 02115, USA atomic force microscopy [9 ]. -

Implications of Metastable Nicks and Nicked Holliday Junctions in Processing Joint Molecules in Mitosis and Meiosis
G C A T T A C G G C A T genes Concept Paper Implications of Metastable Nicks and Nicked Holliday Junctions in Processing Joint Molecules in Mitosis and Meiosis Félix Machín 1,2,3 1 Unidad de Investigación, Hospital Universitario Nuestra Señora de Candelaria, 38010 Santa Cruz de Tenerife, Spain; [email protected] 2 Instituto de Tecnologías Biomédicas, Universidad de la Laguna, 38200 Tenerife, Spain 3 Facultad de Ciencias de la Salud, Universidad Fernando Pessoa Canarias, 35450 Las Palmas de Gran Canaria, Spain Received: 22 October 2020; Accepted: 9 December 2020; Published: 12 December 2020 Abstract: Joint molecules (JMs) are intermediates of homologous recombination (HR). JMs rejoin sister or homolog chromosomes and must be removed timely to allow segregation in anaphase. Current models pinpoint Holliday junctions (HJs) as a central JM. The canonical HJ (cHJ) is a four-way DNA that needs specialized nucleases, a.k.a. resolvases, to resolve into two DNA molecules. Alternatively, a helicase–topoisomerase complex can deal with pairs of cHJs in the dissolution pathway. Aside from cHJs, HJs with a nick at the junction (nicked HJ; nHJ) can be found in vivo and are extremely good substrates for resolvases in vitro. Despite these findings, nHJs have been neglected as intermediates in HR models. Here, I present a conceptual study on the implications of nicks and nHJs in the final steps of HR. I address this from a biophysical, biochemical, topological, and genetic point of view. My conclusion is that they ease the elimination of JMs while giving genetic directionality to the final products. -

The Roles of Hmsh4-Hmsh5 and Hmlh1-Hmlh3 in Meiotic Double Strand Break Repair DISSERTATION Presented in Partial Fulfillment Of
The roles of hMSH4-hMSH5 and hMLH1-hMLH3 in meiotic double strand break repair DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Randal James Soukup Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2016 Dissertation Committee: Richard Fishel, PhD, Advisor Mark Parthun, PhD Charles Bell, PhD Mamuka Kvaratskhelia, PhD Copyrighted by Randal James Soukup 2016 Abstract The DNA double strand break is a highly cytotoxic DNA lesion. Mouse and human mitotically dividing cells experience ~10 double strand breaks (DSBs) per day that are often repaired through non-homologous end joining and result in the accumulation of short insertions and deletions. However, in prophase I of meiosis, ~400 double strand breaks are introduced into primary mouse spermatocytes by the endonuclease SPO11. The cell undergoes a cell-wide DSB repair response which functions to repair each break, and in doing so, pair homologous chromosomes for segregation at the outset of meiosis I. This process generates genetic crossovers between the homologous chromosomes, which are required for accurate chromosome segregation and are also the basis for the reshuffling of genes between maternal and paternal chromosomes. At the center of this DNA repair process is the Holliday Junction, which physically links homologous chromosomes and whose resolution defines the outcome to a genetic crossover or gene conversion event. A number of proteins involved in mitotic DSB repair are also involved with the meiotic process. However, MSH4-MSH5 and MLH1-MLH3 proteins appear to have unique roles in establishing homologous chromosome pairing and segregation during meiotic DSB repair, but do not play any role in mitotic DSB repair. -

Mechanism of Holliday Junction Resolution by the Human GEN1 Protein
Downloaded from genesdev.cshlp.org on October 1, 2021 - Published by Cold Spring Harbor Laboratory Press Mechanism of Holliday junction resolution by the human GEN1 protein Ulrich Rass,1 Sarah A. Compton,2 Joao Matos,1 Martin R. Singleton,3 Stephen C.Y. Ip,1,4 Miguel G. Blanco,1 Jack D. Griffith,2 and Stephen C. West1,5 1London Research Institute, Cancer Research UK, Clare Hall Laboratories, South Mimms, Herts EN6 3LD, United Kingdom; 2Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, North Carolina 27599, USA; 3London Research Institute, Cancer Research UK, London WC2A 3PX, United Kingdom Holliday junction (HJ) resolution is essential for chromosome segregation at meiosis and the repair of stalled/ collapsed replication forks in mitotic cells. All organisms possess nucleases that promote HJ resolution by the introduction of symmetrically related nicks in two strands at, or close to, the junction point. GEN1, a member of the Rad2/XPG nuclease family, was isolated recently from human cells and shown to promote HJ resolution in vitro and in vivo. Here, we provide the first biochemical/structural characterization of GEN1, showing that, like the Escherichia coli HJ resolvase RuvC, it binds specifically to HJs and resolves them by a dual incision mechanism in which nicks are introduced in the pair of continuous (noncrossing) strands within the lifetime of the GEN1–HJ complex. In contrast to RuvC, but like other Rad2/XPG family members such as FEN1, GEN1 is a monomeric 59-flap endonuclease. However, the unique feature of GEN1 that distinguishes it from other Rad2/ XPG nucleases is its ability to dimerize on HJs. -
The Structural Ensemble of a Holliday Junction Determined by X-Ray Scattering Interference Thomas Zettl 1,2, Xuesong Shi2, Steve Bonilla2,3, Steffen M
8090–8098 Nucleic Acids Research, 2020, Vol. 48, No. 14 Published online 29 June 2020 doi: 10.1093/nar/gkaa509 The structural ensemble of a Holliday junction determined by X-ray scattering interference Thomas Zettl 1,2, Xuesong Shi2, Steve Bonilla2,3, Steffen M. Sedlak 1, Jan Lipfert 1,* and Daniel Herschlag2,3,4,5,* 1Department of Physics, Nanosystems Initiative Munich, and Center for Nanoscience, LMU Munich, 80799 Munich, Germany, 2Department of Biochemistry, Stanford University, Stanford, CA 94305, USA, 3Department of Chemical Engineering, Stanford University, Stanford, CA 94305, USA, 4Department of Chemistry, Stanford University, 5 Stanford, CA 94305, USA and Stanford ChEM-H, Stanford University, Stanford, CA 94305, USA Downloaded from https://academic.oup.com/nar/article/48/14/8090/5864709 by guest on 19 August 2020 Received October 13, 2019; Revised May 31, 2020; Editorial Decision June 02, 2020; Accepted June 26, 2020 ABSTRACT other cellular processes (1–4) and have been developed as a widely used tool in DNA nanotechnology (5–7). Holli- The DNA four-way (Holliday) junction is the central in- day junctions, and nucleic acid junctions in general, often termediate of genetic recombination, yet key aspects rely on conformational changes to carry out their biolog- of its conformational and thermodynamic proper- ical functions, either alone or as building blocks of larger ties remain unclear. While multiple experimental ap- nucleic acid and nucleic acid-protein complexes (4,8–13). proaches have been used to characterize the canon- Many proteins, such as junction-resolving enzymes, rec- ical X-shape conformers under specific ionic con- ognize and distort the structure of junctions, for example ditions, the complete conformational ensemble of by stabilizing unstacked conformations, breaking central this motif, especially at low ionic conditions, remains base pairs or changing the inter-duplex angle between the largely undetermined. -

Structural and Biochemical Characterization of Human GEN1 Holliday Junction Resolvase
Dissertation zur Erlangung des Doktorgrades der Fakultät für Chemie und Pharmazie der Ludwigs-Maximilians Universität München Structural and Biochemical Characterization of Human GEN1 Holliday Junction Resolvase Shun-Hsiao Lee aus Hong Kong 2017 ii Erklärung Diese Dissertation wurde im Sinne von § 7 der Promotionsordnung vom 28. November 2011 von Frau Prof. Dr. Elena Conti betreut. Eidesstattliche Versicherung Diese Dissertation wurde eigenständig und ohne unerlaubte Hilfe erarbeitet. 5.12.2017 München, ..................................... ............................................................................. Shun-Hsiao Lee Dissertation eingereicht am 22.09.2017 Erstgutachter: Prof. Dr. Elena Conti Zweitgutachter: Prof. Dr. Karl-Peter Hopfner Mündliche Prüfung am 21.11.2017 iii iv Summary Homologous recombination (HR) is a universal mechanism found in all domains of life for DNA segregation as well as for “error-free” DNA double-strand break (DSB) repair. The key intermediate in this process is a DNA four-way junction also known as a Holliday Junction (HJ), which is formed after pairing of homologous DNA and strand exchange. In later stages of HR, joint DNA molecules have to be faithfully eliminated in order to avoid improper chromosome segregation and to maintain the genome integrity. Human GEN1 is a member of the Rad2/XPG nuclease family and it is the classical Holliday junction-resolving enzyme. Interestingly, this enzyme has no sequence homology to the well-characterized resolvases found in prokaryotes but shares the same function in symmetric HJ cleavage. In contrast to other members of the Rad2/XPG family, GEN1 can form a homodimer and recognize DNA four-way junctions as well as 5’ flaps, replication forks and splayed arms. Its roles in removing recombination and replication intermediates have been implicated in vivo. -

And Mutl-Related Heterocomplexes
Commentary Multiple functions of MutS- and MutL-related heterocomplexes Takuro Nakagawa, Abhijit Datta, and Richard D. Kolodner* Ludwig Institute for Cancer Research, Department of Medicine and Cancer Center, University of California San Diego School of Medicine, CMME 3080, 9500 Gilman Drive, La Jolla, CA 92093-0660 eterodimeric complexes of MutS- paired in msh4, zip1, and mer3 mutants (3, from zygotene (synapsis is started) Hand MutL-related proteins were 9, 14). zip1, zip2, and mer3 mutations also through pachytene (homologs are fully identified from studies of DNA mismatch cause cell-cycle arrest before meiosis I. synapsed) in oocytes (21–23). In contrast, repair (MMR) and have been implicated Although homolog nondisjunction is a pri- MLH1 is detected only at pachytene in in processing of recombination interme- mary cause of missegregation in crossing- spermatocytes. Although human PMS1 is diates. In a recent issue of PNAS, Wang et over-deficient mutants, precocious sepa- most closely related to the yeast MLH3 al. (1) have reported several observations ration of sister chromatids has also been and interacts with MLH1, it is also highly about the function of MutL-related genes observed in msh4, msh5, and zip1 mutants. related to yeast MLH2 (12, 24). Interest- of the yeast Saccharomyces cerevisiae in ZIP1 localizes continuously along the ingly, Pms1Ϫ͞Ϫ mice are fertile, suggesting MMR and recombination in meiosis. entire length of meiotic chromosomes few, if any, meiotic defects (25). Human First, they have observed that MLH1 is the when homologs are synapsed, which de- PMS2 (the yeast PMS1 homolog) forms a common subunit in three different MutL- pends on ZIP2 (7, 8).