Nuclease-Hypersensitive Regions in Chromosomes (Active Genes/Nuclease-Hypersensitive!Cleavage) M
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Endogenous Reverse Transcriptase and Rnase H-Mediated Antiviral Mechanism in Embryonic Stem Cells
www.nature.com/cr www.cell-research.com ARTICLE Endogenous reverse transcriptase and RNase H-mediated antiviral mechanism in embryonic stem cells Junyu Wu1, Chunyan Wu1, Fan Xing1, Liu Cao1, Weijie Zeng1, Liping Guo1, Ping Li1, Yongheng Zhong1, Hualian Jiang1, Manhui Luo1, Guang Shi2, Lang Bu1, Yanxi Ji1, Panpan Hou1, Hong Peng1, Junjiu Huang2, Chunmei Li1 and Deyin Guo 1 Nucleic acid-based systems play important roles in antiviral defense, including CRISPR/Cas that adopts RNA-guided DNA cleavage to prevent DNA phage infection and RNA interference (RNAi) that employs RNA-guided RNA cleavage to defend against RNA virus infection. Here, we report a novel type of nucleic acid-based antiviral system that exists in mouse embryonic stem cells (mESCs), which suppresses RNA virus infection by DNA-mediated RNA cleavage. We found that the viral RNA of encephalomyocarditis virus can be reverse transcribed into complementary DNA (vcDNA) by the reverse transcriptase (RTase) encoded by endogenous retrovirus-like elements in mESCs. The vcDNA is negative-sense single-stranded and forms DNA/RNA hybrid with viral RNA. The viral RNA in the heteroduplex is subsequently destroyed by cellular RNase H1, leading to robust suppression of viral growth. Furthermore, either inhibition of the RTase activity or depletion of endogenous RNase H1 results in the promotion of virus proliferation. Altogether, our results provide intriguing insights into the antiviral mechanism of mESCs and the antiviral function of endogenized retroviruses and cellular RNase H. Such a natural nucleic acid-based antiviral mechanism in mESCs is referred to as ERASE (endogenous RTase/RNase H-mediated antiviral system), which is an addition to the previously known nucleic acid-based antiviral mechanisms including CRISPR/Cas in bacteria and RNAi in plants and invertebrates. -

Specific Principles of Genome-Wide RNA-Chromatin Interactions
ARTICLE There are amendments to this paper https://doi.org/10.1038/s41467-020-14337-6 OPEN RADICL-seq identifies general and cell type–specific principles of genome-wide RNA-chromatin interactions Alessandro Bonetti 1,2,18*, Federico Agostini 3,18, Ana Maria Suzuki1,4, Kosuke Hashimoto1, Giovanni Pascarella1, Juliette Gimenez 5, Leonie Roos6,7, Alex J. Nash6,7, Marco Ghilotti1, Christopher J. F. Cameron8,9, Matthew Valentine 1, Yulia A. Medvedeva10,11,12, Shuhei Noguchi1, Eneritz Agirre 2, Kaori Kashi1, Samudyata2, Joachim Luginbühl1, Riccardo Cazzoli13, Saumya Agrawal1, Nicholas M. Luscombe 3,14,15, Mathieu Blanchette8, Takeya Kasukawa 1, Michiel de Hoon1, Erik Arner1, 1234567890():,; Boris Lenhard 6,7,16, Charles Plessy 1, Gonçalo Castelo-Branco 2, Valerio Orlando5,17* & Piero Carninci 1* Mammalian genomes encode tens of thousands of noncoding RNAs. Most noncoding tran- scripts exhibit nuclear localization and several have been shown to play a role in the reg- ulation of gene expression and chromatin remodeling. To investigate the function of such RNAs, methods to massively map the genomic interacting sites of multiple transcripts have been developed; however, these methods have some limitations. Here, we introduce RNA And DNA Interacting Complexes Ligated and sequenced (RADICL-seq), a technology that maps genome-wide RNA–chromatin interactions in intact nuclei. RADICL-seq is a proximity ligation-based methodology that reduces the bias for nascent transcription, while increasing genomic coverage and unique mapping rate efficiency compared with existing methods. RADICL-seq identifies distinct patterns of genome occupancy for different classes of tran- scripts as well as cell type–specific RNA-chromatin interactions, and highlights the role of transcription in the establishment of chromatin structure. -

(NAP) and Illustration of DNA Flexure Angles at Single Molecule Resolution Debayan Purkait†A, Debolina Bandyopadhyay†A, and Padmaja P
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.11.293639; this version posted September 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Vital Insights into Prokaryotic Genome Compaction by Nucleoid-Associated Protein (NAP) and Illustration of DNA Flexure Angles at Single Molecule Resolution Debayan Purkait†a, Debolina Bandyopadhyay†a, and Padmaja P. Mishra*a aChemical Sciences Division, Saha Institute of Nuclear Physics, India aHomi Bhaba National Institute (HBNI), India †Both the authors have contributed equally. * Corresponding author Abstract Integration Host Factor (IHF) is a heterodimeric site-specific nucleoid-associated protein (NAP) well known for its DNA bending ability. The binding is mediated through the narrow minor grooves of the consensus sequence, involving van der-Waals interaction and hydrogen bonding. Although the DNA bend state of IHF has been captured by both X-ray Crystallography and Atomic Force Microscopy (AFM), the range of flexibility and degree of heterogeneity in terms of quantitative analysis of the nucleoprotein complex has largely remained unexplored. Here we have monitored and compared the trajectories of the conformational dynamics of a dsDNA upon binding of wild-type (wt) and single-chain (sc) IHF at millisecond resolution through single-molecule FRET (smFRET). Our findings reveal that the nucleoprotein complex exists in a ‘Slacked-Dynamic’ state throughout the observation window where many of them have switched between multiple ‘Wobbling States’ in the course of attainment of packaged form. A range of DNA ‘Flexure Angles’ has been calculated that give us vital insights regarding the nucleoid organization and transcriptional regulation in prokaryotes. -

Cell Cycle Regulation by the Bacterial Nucleoid
Available online at www.sciencedirect.com ScienceDirect Cell cycle regulation by the bacterial nucleoid David William Adams, Ling Juan Wu and Jeff Errington Division site selection presents a fundamental challenge to all terminate in the terminus region (Ter; 1808). Once organisms. Bacterial cells are small and the chromosome chromosome replication and segregation are complete (nucleoid) often fills most of the cell volume. Thus, in order to the cell is ready to divide. In Bacteria this normally occurs maximise fitness and avoid damaging the genetic material, cell by binary fission and in almost all species this is initiated division must be tightly co-ordinated with chromosome by the assembly of the tubulin homologue FtsZ into a replication and segregation. To achieve this, bacteria employ a ring-like structure (‘Z-ring’) at the nascent division site number of different mechanisms to regulate division site (Figure 1) [1]. The Z-ring then functions as a dynamic selection. One such mechanism, termed nucleoid occlusion, platform for assembly of the division machinery [2,3]. Its allows the nucleoid to protect itself by acting as a template for central role in division also allows FtsZ to serve as a nucleoid occlusion factors, which prevent Z-ring assembly over regulatory hub for the majority of regulatory proteins the DNA. These factors are sequence-specific DNA-binding identified to date [2,4]. Nevertheless, the precise ultra- proteins that exploit the precise organisation of the nucleoid, structure of the Z-ring and whether or not it plays a direct allowing them to act as both spatial and temporal regulators of role in force-generation during division remains contro- bacterial cell division. -

Pharmacokinetics, Pharmacodynamics and Metabolism Of
PHARMACOKINETICS, PHARMACODYNAMICS AND METABOLISM OF GTI-2040, A PHOSPHOROTHIOATE OLIGONUCLEOTIDE TARGETING R2 SUBUNIT OF RIBONUCLEOTIDE REDUCTASE DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Xiaohui Wei, M.S. * * * * * * The Ohio State University 2006 Approved by Dissertation Committee: Dr. Kenneth K. Chan, Adviser Adviser Dr. Guido Marcucci, Co-adviser Graduate Program in Pharmacy Dr. Thomas D. Schmittgen Dr. Robert J. Lee Co-Adviser Graduate Program in Pharmacy ABSTRACT Over the last several decades, antisense therapy has been developed into a promising gene-targeted strategy to specifically inhibit the gene expression. Ribonucleotide reductase (RNR), composing of subunits R1 and R2, is an important enzyme involved in the synthesis of all of the precursors used in DNA replication. Over- expression of R2 has been found in almost every type of cancer studied. GTI-2040 is a 20-mer phosphorothioate oligonucleotide targeting the coding region in mRNA of the R2 component of human RNR. In this project, clinical pharamcokinetics (PK), pharmacodynamics (PD) and metabolism of this novel therapeutics were investigated in patients with acute myeloid leukemia (AML). A picomolar specific hybridization-ligation ELISA method has been developed and validated for quantification of GTI-2040. GTI-2040 and neophectin complex was found to enhance drug cellular uptake and exhibited sequence- and dose-dependent down-regulation of R2 mRNA and protein in K562 cells. Robust intracellular concentrations (ICs) of GTI-2040 were achieved in peripheral blood mononuclear cells (PBMC) and bone marrow (BM) cells from treated AML patients. GTI-2040 concentrations in the nucleus of BM cells were found to correlate with the R2 mRNA down-regulation and disease response. -

Catalytic Domain of Plasmid Pad1 Relaxase Trax Defines a Group Of
Catalytic domain of plasmid pAD1 relaxase TraX defines a group of relaxases related to restriction endonucleases María Victoria Franciaa,1, Don B. Clewellb,c, Fernando de la Cruzd, and Gabriel Moncaliánd aServicio de Microbiología, Hospital Universitario Marqués de Valdecilla e Instituto de Formación e Investigación Marqués de Valdecilla, Santander 39008, Spain; bDepartment of Biologic and Materials Sciences, University of Michigan School of Dentistry, Ann Arbor, MI 48109; cDepartment of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, MI 48109; and dDepartamento de Biología Molecular e Instituto de Biomedicina y Biotecnología de Cantabria, Universidad de Cantabria–Consejo Superior de Investigaciones Científicas–Sociedad para el Desarrollo Regional de Cantabria, Santander 39011, Spain Edited by Roy Curtiss III, Arizona State University, Tempe, AZ, and approved July 9, 2013 (received for review May 30, 2013) Plasmid pAD1 is a 60-kb conjugative element commonly found in known relaxases show two characteristic sequence motifs, motif I clinical isolates of Enterococcus faecalis. The relaxase TraX and the containing the catalytic Tyr residue (which covalently attaches to primary origin of transfer oriT2 are located close to each other and the 5′ end of the cleaved DNA) and motif III with the His-triad have been shown to be essential for conjugation. The oriT2 site essential for relaxase activity (facilitating the cleavage reaction contains a large inverted repeat (where the nic site is located) by activation of the catalytic Tyr). This His-triad has been used as adjacent to a series of short direct repeats. TraX does not show a relaxase diagnostic signature (12). fi any of the typical relaxase sequence motifs but is the prototype of Plasmid pAD1 relaxase, TraX, was identi ed (9) and shown to oriT2 a unique family of relaxases (MOB ). -

Relaxation Complexes of Plasmids Colel and Cole2:Unique Site of The
Proc. Nat. Acad. Sci. USA Vol. 71, No. 10, pp. 3854-3857, October 1974 Relaxation Complexes of Plasmids ColEl and ColE2: Unique Site of the Nick in the Open Circular DNA of the Relaxed Complexes (Escherichia coli/supercoiled DNA/endonuclease/restriction enzyme/DNA replication) MICHAEL A. LOVETT, DONALD G. GUINEY, AND DONALD R. HELINSKI Department of Biology, University of California, San Diego, La Jolla, Calif. 92037 Communicated by Stanley L. Miller, June 26, 1974 ABSTRACT The product of the induced relaxation of volved in the production of a single-strand cleavage in the supercoiled DNA-protein relaxation complexes of colicino- initiation of replication and/or conjugal transfer of plasmid genic factors El (ColEl) and E2 (CoIE2) is an open circular DNA molecule with a strand-specific nick. Cleavage of the DNA. An involvement in such processes would most likely open circular DNA of each relaxed complex with the EcoRI require that the relaxation event take place at a unique site restriction endonuclease demonstrates that the single- in the DNA molecule. In this report, evidence will be pre- strand break is at a unique position. The site of the single- sented from studies using the EcoRI restriction endonuclease strand break in the relaxed ColEl complex is approximately the same distance from the EcoRI cleavage site as the that the strand-specific relaxation event takes place at a origin of ColEl DNA replication. unique site on both the ColEl and CoIE2 plasmid molecules. The location of this site, at least in the case of the ColEl Many of extrachromosomal circular DNA elements (plas- plasmid, is approximately the same distance from the single mids) of Escherichia coli, differing in molecular weight, num- EcoRI site as the origin of replication for this plasmid. -

Termin Translat Trna Utr Mutat Protein Signal
Drugs & Chemicals 1: Tumor Suppressor Protein p53 2: Heterogeneous-Nuclear Ribonucleo- (1029) proteins (14) activ apoptosi arf cell express function inactiv induc altern assai associ bind mdm2 mutat p53 p73 pathwai protein regul complex detect exon famili genom respons suppress suppressor tumor wild-typ interact intron isoform nuclear protein sensit site specif splice suggest variant 3: RNA, Transfer (110) 4: DNA Primers (1987) codon contain differ eukaryot gene initi amplifi analysi chain clone detect dna express mrna protein region ribosom rna fragment gene genotyp mutat pcr sequenc site speci suggest synthesi polymorph popul primer reaction region restrict sequenc speci termin translat trna utr 5: Saccharomyces cerevisiae Proteins 6: Apoptosis Regulatory Proteins (291) (733) activ apoptosi apoptosis-induc albican bud candida cerevisia complex encod apoptot bcl-2 caspas caspase-8 cell eukaryot fission function growth interact involv death fasl induc induct ligand methyl necrosi pathwai program sensit surviv trail mutant pomb protein requir saccharomyc strain suggest yeast 7: Plant Proteins (414) 8: Membrane Proteins (1608) access arabidopsi cultivar flower hybrid leaf leav apoptosi cell conserv domain express function gene human identifi inhibitor line maiz plant pollen rice root seed mammalian membran mice mous mutant seedl speci thaliana tomato transgen wheat mutat protein signal suggest transport 1 9: Tumor Suppressor Proteins (815) 10: 1-Phosphatidylinositol 3-Kinase activ arrest cell cycl cyclin damag delet dna (441) 3-kinas activ -
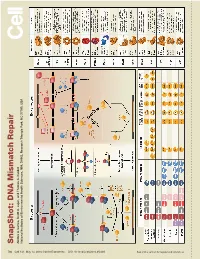
S Na P S H O T: D N a Mism a Tc H R E P a Ir
SnapShot: Repair DNA Mismatch Scott A. Lujan, and Thomas Kunkel A. Larrea, Andres Park, NC 27709, USA Triangle Health Sciences, NIH, DHHS, Research National Institutes of Environmental 730 Cell 141, May 14, 2010 ©2010 Elsevier Inc. DOI 10.1016/j.cell.2010.05.002 See online version for legend and references. SnapShot: DNA Mismatch Repair Andres A. Larrea, Scott A. Lujan, and Thomas A. Kunkel National Institutes of Environmental Health Sciences, NIH, DHHS, Research Triangle Park, NC 27709, USA Mismatch Repair in Bacteria and Eukaryotes Mismatch repair in the bacterium Escherichia coli is initiated when a homodimer of MutS binds as an asymmetric clamp to DNA containing a variety of base-base and insertion-deletion mismatches. The MutL homodimer then couples MutS recognition to the signal that distinguishes between the template and nascent DNA strands. In E. coli, the lack of adenine methylation, catalyzed by the DNA adenine methyltransferase (Dam) in newly synthesized GATC sequences, allows E. coli MutH to cleave the nascent strand. The resulting nick is used for mismatch removal involving the UvrD helicase, single-strand DNA-binding protein (SSB), and excision by single-stranded DNA exonucleases from either direction, depending upon the polarity of the nick relative to the mismatch. DNA polymerase III correctly resynthesizes DNA and ligase completes repair. In bacteria lacking Dam/MutH, as in eukaryotes, the signal for strand discrimination is uncertain but may be the DNA ends associated with replication forks. In these bacteria, MutL harbors a nick-dependent endonuclease that creates a nick that can be used for mismatch excision. Eukaryotic mismatch repair is similar, although it involves several dif- ferent MutS and MutL homologs: MutSα (MSH2/MSH6) recognizes single base-base mismatches and 1–2 base insertion/deletions; MutSβ (MSH2/MSH3) recognizes insertion/ deletion mismatches containing two or more extra bases. -

Supplementary Table S1 List of Proteins Identified with LC-MS/MS in the Exudates of Ustilaginoidea Virens Mol
Supplementary Table S1 List of proteins identified with LC-MS/MS in the exudates of Ustilaginoidea virens Mol. weight NO a Protein IDs b Protein names c Score d Cov f MS/MS Peptide sequence g [kDa] e Succinate dehydrogenase [ubiquinone] 1 KDB17818.1 6.282 30.486 4.1 TGPMILDALVR iron-sulfur subunit, mitochondrial 2 KDB18023.1 3-ketoacyl-CoA thiolase, peroxisomal 6.2998 43.626 2.1 ALDLAGISR 3 KDB12646.1 ATP phosphoribosyltransferase 25.709 34.047 17.6 AIDTVVQSTAVLVQSR EIALVMDELSR SSTNTDMVDLIASR VGASDILVLDIHNTR 4 KDB11684.1 Bifunctional purine biosynthetic protein ADE1 22.54 86.534 4.5 GLAHITGGGLIENVPR SLLPVLGEIK TVGESLLTPTR 5 KDB16707.1 Proteasomal ubiquitin receptor ADRM1 12.204 42.367 4.3 GSGSGGAGPDATGGDVR 6 KDB15928.1 Cytochrome b2, mitochondrial 34.9 58.379 9.4 EFDPVHPSDTLR GVQTVEDVLR MLTGADVAQHSDAK SGIEVLAETMPVLR 7 KDB12275.1 Aspartate 1-decarboxylase 11.724 112.62 3.6 GLILTLSEIPEASK TAAIAGLGSGNIIGIPVDNAAR 8 KDB15972.1 Glucosidase 2 subunit beta 7.3902 64.984 3.2 IDPLSPQQLLPASGLAPGR AAGLALGALDDRPLDGR AIPIEVLPLAAPDVLAR AVDDHLLPSYR GGGACLLQEK 9 KDB15004.1 Ribose-5-phosphate isomerase 70.089 32.491 32.6 GPAFHAR KLIAVADSR LIAVADSR MTFFPTGSQSK YVGIGSGSTVVHVVDAIASK 10 KDB18474.1 D-arabinitol dehydrogenase 1 19.425 25.025 19.2 ENPEAQFDQLKK ILEDAIHYVR NLNWVDATLLEPASCACHGLEK 11 KDB18473.1 D-arabinitol dehydrogenase 1 11.481 10.294 36.6 FPLIPGHETVGVIAAVGK VAADNSELCNECFYCR 12 KDB15780.1 Cyanovirin-N homolog 85.42 11.188 31.7 QVINLDER TASNVQLQGSQLTAELATLSGEPR GAATAAHEAYK IELELEK KEEGDSTEKPAEETK LGGELTVDER NATDVAQTDLTPTHPIR 13 KDB14501.1 14-3-3 -

Effects of Abasic Sites and DNA Single-Strand Breaks on Prokaryotic RNA Polymerases (Apurinic/Apyrimidinic Sites/Transcription/DNA Damage) WEI ZHOU* and PAUL W
Proc. Natl. Acad. Sci. USA Vol. 90, pp. 6601-6605, July 1993 Biochemistry Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases (apurinic/apyrimidinic sites/transcription/DNA damage) WEI ZHOU* AND PAUL W. DOETSCH*tt Departments of *Biochemistry and tRadiation Oncology and tDivision of Cancer Biology, Emory University School of Medicine, Atlanta, GA 30322 Communicated by Philip C. Hanawalt, April 19, 1993 ABSTRACT Abasic sites are thought to be the most fre- effects of such damage on both transcription initiation and quently occurring cellular DNA damage and are generated elongation processes (19, 20). We wished to determine the spontaneously or as the result of chemical or radiation damage effects of AP sites on the transcription elongation process by to DNA. In contrast to the wealth of information that exists on using a defined system that would provide direct information the effects of abasic sites on DNA polymerases, very little is regarding the ability of RNA polymerases to bypass or be known about how these lesions interact with RNA polymerases. blocked by AP sites and, if bypass was observed, to deter- An in vitro transcription system was used to determine the mine the nature of the base insertion opposite such lesions. effects of abasic sites and single-strand breaks on transcrip- For these studies, we constructed a DNA template containing tional elongation. DNA templates were constructed containing a single AP site placed at a unique location downstream from single abasic sites or nicks placed at unique locations down- either the SP6 or tac promoter and carried out in vitro stream from two different promoters and were transcribed by transcription experiments with SP6 and E. -

Subject Index Proc
13088 Subject Index Proc. Natl. Acad. Sci. USA 91 (1994) Apoptosis in substantia nigra following developmental striatal excitotoxic Brine shrimp injury, 8117 See Artemia Visualizing hippocampal synaptic function by optical detection of Ca2l Broccol entry through the N-methyl-D-aspartate channel, 8170 See Brassica Amygdala modulation of hippocampal-dependent and caudate Bromophenacyl bromide nucleus-dependent memory processes, 8477 Bromophenacyl bromide binding to the actin-bundling protein I-plastin Distribution of corticotropin-releasing factor receptor mRNA expression inhibits inositol trisphosphate-independent increase in Ca2l in human in the rat brain and pituitary, 8777 neutrophils, 3534 Brownian dynamics Preproenkephalin promoter yields region-specific and long-term Adhesion of hard spheres under the influence of double-layer, van der expression in adult brain after direct in vivo gene transfer via a Waals, and gravitational potentials at a solid/liquid interface, 3004 defective herpes simplex viral vector, 8979 Browsers Intravenous administration of a transferrin receptor antibody-nerve Thorn-like prickles and heterophylly in Cyanea: Adaptations to extinct growth factor conjugate prevents the degeneration of cholinergic avian browsers on Hawaii?, 2810 striatal neurons in a model of Huntington disease, 9077 Bruton agammaglobulinemia Axotomy induces the expression of vasopressin receptors in cranial and Genomic organization and structure of Bruton agammaglobulinemia spinal motor nuclei in the adult rat, 9636 tyrosine kinase: Localization