Relationships Among Peach, Almond, and Related Species As Detected by Simple Sequence Repeat Markers
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Report of a Working Group on Prunus: Sixth and Seventh Meetings
European Cooperative Programme for Plant Genetic Report of a Working Resources ECP GR Group on Prunus Sixth Meeting, 20-21 June 2003, Budapest, Hungary Seventh Meeting, 1-3 December 2005, Larnaca, Cyprus L. Maggioni and E. Lipman, compilers IPGRI and INIBAP operate under the name Bioversity International Supported by the CGIAR European Cooperative Programme for Plant Genetic Report of a Working Resources ECP GR Group on Prunus Sixth Meeting, 20 –21 June 2003, Budapest, Hungary Seventh Meeting, 1 –3 December 2005, Larnaca, Cyprus L. Maggioni and E. Lipman, compilers ii REPORT OF A WORKING GROUP ON PRUNUS: SIXTH AND SEVENTH MEETINGS Bioversity International is an independent international scientific organization that seeks to improve the well- being of present and future generations of people by enhancing conservation and the deployment of agricultural biodiversity on farms and in forests. It is one of 15 centres supported by the Consultative Group on International Agricultural Research (CGIAR), an association of public and private members who support efforts to mobilize cutting-edge science to reduce hunger and poverty, improve human nutrition and health, and protect the environment. Bioversity has its headquarters in Maccarese, near Rome, Italy, with offices in more than 20 other countries worldwide. The Institute operates through four programmes: Diversity for Livelihoods, Understanding and Managing Biodiversity, Global Partnerships, and Commodities for Livelihoods. The international status of Bioversity is conferred under an Establishment Agreement which, by January 2006, had been signed by the Governments of Algeria, Australia, Belgium, Benin, Bolivia, Brazil, Burkina Faso, Cameroon, Chile, China, Congo, Costa Rica, Côte d’Ivoire, Cyprus, Czech Republic, Denmark, Ecuador, Egypt, Greece, Guinea, Hungary, India, Indonesia, Iran, Israel, Italy, Jordan, Kenya, Malaysia, Mali, Mauritania, Morocco, Norway, Pakistan, Panama, Peru, Poland, Portugal, Romania, Russia, Senegal, Slovakia, Sudan, Switzerland, Syria, Tunisia, Turkey, Uganda and Ukraine. -

Factores Moleculares Implicados En El Sistema De Incompatibilidad Floral En Almendro [Prunus Dulcis (Miller) D
UNIVERSIDAD DE MURCIA FACULTAD DE BIOLOGÍA Factores Moleculares Implicados en el Sistema de Incompatibilidad Floral en Almendro [Prunus dulcis (Miller) D. A. Webb] Dª Eva María Gómez González 2017 TESIS DOCTORAL “Factores moleculares implicados en el sistema de incompatibilidad floral en almendro [Prunus dulcis (Miller) D.A. Webb]” Doctoranda: Eva María Gómez González Directores: Dra. Encarnación Ortega Pastor Dr. Federico Dicenta López-Higuera Murcia, 2017 CENTRO DE EDAFOLOGÍA Y BIOLOGÍA APLICADA DEL SEGURA (CEBAS) D. Federico Dicenta López-Higuera, Doctor por la Universidad de Murcia y Dª Encarnación Ortega Pastor, Doctora por la Universidad de Murcia, ambos adscritos al Centro de Edafología y Biología Aplicada del Segura, del Consejo Superior de Investigaciones Científicas. AUTORIZAN: La presentación de la Tesis Doctoral titulada “Factores moleculares implicados en el sistema de incompatibilidad floral en almendro [Prunus dulcis (Miller) D.A. Webb”, realizada por Dª. Eva María Gómez González, Licenciada en Bioquímica, bajo nuestra inmediata dirección y supervisión, en el Departamento de Mejora Vegetal del Centro de Edafología y Biología Aplicada del Segura (CEBAS-CSIC) de Murcia. Considerando que se trata de un trabajo original de investigación que reúne los requisitos establecidos en el RD 1393/2007, de 29 de octubre, estimamos que puede ser presentado para la obtención del grado de Doctor por la Universidad de Murcia. Murcia, 31 de Mayo de 2017 Campus Universitario de Espinardo 30100 Espinardo, Murcia. ESPAÑA Telf. (34) 968 396200 Fax.: (34) 968 396213 Agradecimientos La realización de esta tesis doctoral ha sido posible gracias al disfrute de la beca FPI (BES-2011-050500), asociada al proyecto “Mejora genética del Almendro” (AGL2010-22197-C02-02) financiado por Ministerio de Economía y Competitividad. -

Nursery Price List
Lincoln-Oakes Nurseries 3310 University Drive • Bismarck, ND 58504 Nursery Seed Price List 701-223-8575 • [email protected] The following seed is in stock or will be collected and available for 2010 or spring 2011 PENDING CROP, all climatic zone 3/4 collections from established plants in North Dakota except where noted. Acer ginnala - 18.00/lb d.w Cornus racemosa - 19.00/lb Amur Maple Gray dogwood Acer tataricum - 15.00/lb d.w Cornus alternifolia - 21.00/lb Tatarian Maple Pagoda dogwood Aesculus glabra (ND, NE) - 3.95/lb Cornus stolonifera (sericea) - 30.00/lb Ohio Buckeye – collected from large well performing Redosier dogwood Trees in upper midwest Amorpha canescens - 90.00/lb Leadplant 7.50/oz Amorpha fruiticosa - 10.50/lb False Indigo – native wetland restoration shrub Aronia melanocarpa ‘McKenzie” - 52.00/lb Black chokeberry - taller form reaching 6-8 ft in height, glossy foliage, heavy fruit production, Corylus cornuta (partial husks) - 16.00/lb NRCS release Beaked hazelnut/Native hazelnut (Inquire) Caragana arborescens - 16.00/lb Cotoneaster integerrimus ‘Centennial’ - 32.00/lb Siberian peashrub European cotoneaster – NRCS release, 6-10’ in height, bright red fruit Celastrus scandens (true) (Inquire) - 58.00/lb American bittersweet, no other contaminating species in area Crataegus crus-galli - 22.00/lb Cockspur hawthorn, seed from inermis Crataegus mollis ‘Homestead’ arnoldiana-24.00/lb Arnold hawthorn – NRCS release Crataegus mollis - 19.50/lb Downy hawthorn Elaeagnus angustifolia - 9.00/lb Russian olive Elaeagnus commutata -

Plant Species Richness and Composition of a Habitat Island
Biodiversity Data Journal 8: e48704 doi: 10.3897/BDJ.8.e48704 Research Article Plant species richness and composition of a habitat island within Lake Kastoria and comparison with those of a true island within the protected Pamvotis lake (NW Greece) Alexandros Papanikolaou‡‡, Maria Panitsa ‡ Division of Plant Biology, Department of Biology, University of Patras, Patras, Greece Corresponding author: Maria Panitsa ([email protected]) Academic editor: Gianniantonio Domina Received: 22 Nov 2019 | Accepted: 07 Jan 2020 | Published: 15 Jan 2020 Citation: Papanikolaou A, Panitsa M (2020) Plant species richness and composition of a habitat island within Lake Kastoria and comparison with those of a true island within the protected Pamvotis lake (NW Greece). Biodiversity Data Journal 8: e48704. https://doi.org/10.3897/BDJ.8.e48704 Abstract Lake Kastoria is one of the potentially “ancient” Balkan lakes that has a great environmental importance and ecological value, attracts high touristic interest and is under various anthropogenic pressures. It belongs to a Natura 2000 Special Protection Area and a Site of Community Interest. The city of Kastoria is located at the western part of the lake and just next to it, towards the centre of the lake, is a peninsula, a habitat island. In the framework of research concerning the flora of lake islands of Greece, one of the main objectives of the present study is to fill a gap concerning plant species richness of the habitat island within the protected Lake Kastoria, which is surrounded by the lake except for its north-western part where the border of the city of Kastoria is located. -

The Ornamental Trees of South Dakota N.E
South Dakota State University Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange South Dakota State University Agricultural Bulletins Experiment Station 4-1-1931 The Ornamental Trees of South Dakota N.E. Hansen Follow this and additional works at: http://openprairie.sdstate.edu/agexperimentsta_bulletins Recommended Citation Hansen, N.E., "The Ornamental Trees of South Dakota" (1931). Bulletins. Paper 260. http://openprairie.sdstate.edu/agexperimentsta_bulletins/260 This Bulletin is brought to you for free and open access by the South Dakota State University Agricultural Experiment Station at Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange. It has been accepted for inclusion in Bulletins by an authorized administrator of Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange. For more information, please contact [email protected]. Bulletin 260 April, 1931 The Ornamental Trees of South Dakota Figure I-The May Day Tree. Horticulture Department Agricultural Experiment Station South Dakota State College of Agriculture and Mechanic Arts Brookings, S. Dak. The Ornamental Trees of South Dakota N. E. Hansen This bulletin describes the deciduous trees. By deciduous trees is meant those that shed their leaves in winter. The evergreens of South Dakota are described in bulletin 254, October 1930. A bulletin on "The Ornamental Shrubs of South Dakota" is ready for early publication. The following list should be studied in connection with the trees described in South Dakota bulletin 246, "'The Shade, Windbreak and Timber Trees of South Dakota," 48 pages, March 1930. All the trees in both bulletins have ornamental value in greater or less degree. -

Seed Germination and Seedling Establishment of Some Wild Almond Species
African Journal of Biotechnology Vol. 10(40), pp. 7780-7786, 1 August, 2011 Available online at http://www.academicjournals.org/AJB DOI: 10.5897/AJB10.1064 ISSN 1684–5315 © 2011 Academic Journals Full Length Research Paper Seed germination and seedling establishment of some wild almond species Alireza Rahemi 1* , Toktam Taghavi 2, Reza Fatahi 2, Ali Ebadi 2, Darab Hassani 3, José Chaparro 4 and Thomas Gradziel 5 1Department of Horticultural Science, Azad University (Science and Research Branch), Tehran, Iran. 2Department of Horticultural Science, University of Tehran, Karaj, Iran. 3Department of Horticulture, Seed and Plant Improvement Institute, Karaj, Iran. 4Department of Horticultural Science, University of Florida, Gainesville, USA. 5 Department of Plant Sciences, University of California, Davis. USA. Accepted 20 January, 2011 Wild almond species are important genetic resources for resistance to unsuitable condition, especially drought stress. They have been used traditionally as rootstocks in some areas of Iran. So far, 21 wild almond species and 7 inter species hybrids have been identified in Iran. To study seed germination and seedling establishment of some of these species, three separate experiments were designed. In the first experiment, the application of gibberellic acid (GA3) (0, 250, 500 and 750 ppm) for 24 h was studied on germination characteristics of four wild almond accessions after stratification at 5 ± 0.5°C in Perlite media. Germination percentage, index vigor and root initiation factors were different in almond accessions, but were not affected by hormonal treatments. In the second experiment, seeds of another six wild almond accessions were stratified to compare their germination ability. Germination percentage, index vigor and root initiation were different among accessions significantly. -

Gene Flow in Prunus Species in the Context of Novel Trait Risk Assessment
Environ. Biosafety Res. 9 (2010) 75–85 Available online at: c ISBR, EDP Sciences, 2011 www.ebr-journal.org DOI: 10.1051/ebr/2010011 Gene flow in Prunus species in the context of novel trait risk assessment S. Zahra H. Cici* and Rene C. Van Acker1 Department of Plant Agriculture, University of Guelph, Guelph, ON, N1G2W1, Canada Prunus species are important commercial fruit (plums, apricot, peach and cherries), nut (almond) and orna- mental trees cultivated broadly worldwide. This review compiles information from available literature on Prunus species in regard to gene flow and hybridization within this complex of species. The review serves as a resource for environmental risk assessment related to pollen mediated gene flow and the release of transgenic Prunus. It reveals that Prunus species, especially plums and cherries show high potential for transgene flow. A range of characteristics including; genetic diversity, genetic bridging capacity, inter- and intra-specific genetic compat- ibility, self sterility (in most species), high frequency of open pollination, insect assisted pollination, perennial nature, complex phenotypic architecture (canopy height, heterogeneous crown, number of flowers produced in an individual plant), tendency to escape from cultivation, and the existence of ornamental and road side Prunus species suggest that there is a tremendous and complicated ability for pollen mediated gene movement among Prunus species. Ploidy differences among Prunus species do not necessarily provide genetic segregation. The characteristics of Prunus species highlight the complexity of maintaining coexistence between GM and non-GM Prunus if there were commercial production of GM Prunus species. The results of this review suggest that the commercialization of one GM Prunus species can create coexistence issues for commercial non-GM Prunus production. -

175 Section 6 Stone Fruits (Prunus Spp.)
SECTION 6 STONE FRUITS (PRUNUS SPP.) 1. Introduction A. General Background The genus of Prunus sensu latu comprises more domesticated (also cultivated) species of temperate fruits than the other genera in the family of Rosaceae (Malus, Pyrus, Sorbus, Cydonia, Rubus, Fragaria). One of the obvious reasons for the abundant domestication might have been the coincidence between the location of the centre of variability of Prunus and the site of human evolution and/or of the first ancient high civilisations of human history. Improvement of fruit trees through traditional breeding methods is a long-term effort because of their lengthy generation time. Thus, new approaches are researched to attain the envisaged breeding goals in a reasonable time frame. Genetic transformation is potentially useful, because specific genetic changes can be made. In the last few years successful examples of resistance breeding against viruses from different plant virus families have been reported, using the coat protein-mediated cross protection approach (Beachy et al., 1990). However, only very few fruit trees have been among these experiments due to the difficulties in transformation protocols. “Cross protection” was originally described as the phenomenon of protection of a plant against the invasion of a severe disease-causing virus due to prior inoculation of the plant with an attenuated virus strain (McKinney, 1929). Hamilton postulated in 1980 that the expression of sequences from the viral genome, if expressed in transgenic plants, could possibly cause a protection against viruses. In fact by the expression of the viral coat protein gene in transgenic plants, similar effects could be obtained, and it was therefore distinguished as coat protein mediated protection (Beachy et al., 1990). -

Halász Júlia
Doktori (PhD) értekezés A KAJSZI ÖNMEDD İSÉGÉT MEGHATÁROZÓ S-ALLÉL-RENDSZER MOLEKULÁRIS HÁTTERE Halász Júlia Témavezet ı: Dr. Pedryc Andrzej, CSc egyetemi docens Budapesti Corvinus Egyetem Genetika és Növénynemesítés Tanszék Budapest 2007 A doktori iskola megnevezése: Kertészettudományi Doktori Iskola tudományága : Biológiai tudományok vezet ıje: Dr. Papp János, DSc egyetemi tanár BCE, Kertészettudományi Kar, Gyümölcsterm ı Növények Tanszék Témavezet ı: Dr. Pedryc Andrzej, CSc egyetemi docens BCE, Kertészettudományi Kar Genetika és Növénynemesítés Tanszék A jelölt a Budapesti Corvinus Egyetem Doktori Szabályzatában el ıírt valamennyi feltételnek eleget tett, az értekezés m őhelyvitájában elhangzott észrevételeket és javaslatokat az értekezés átdolgozásakor figyelembe vette, ezért az értekezés nyilvános vitára bocsátható. ........................................................... ........................................................... Dr. Papp János Dr. Pedryc Andrzej Az iskolavezet ı jóváhagyása A témavezet ı jóváhagyása A Budapesti Corvinus Egyetem Élettudományi Területi Doktori Tanács 2007. február 13-i határozatában a nyilvános vita lefolytatására az alábbi bíráló Bizottságot jelölte ki: BÍRÁLÓ BIZOTTSÁG : Elnöke Tóth Magdolna, CSc Tagjai Jenes Barnabás, CSc Palkovics László, DSc Szegedi Ern ı, DSc Opponensek Janda Tibor, CSc Kiss Erzsébet, CSc Titkár Kocsisné Molnár Gitta, PhD „A gyümölcsfák életének legérdekesebb részei azok a jelenségek, amelyek ivaros szaporodásával vannak egybekapcsolva. Nagyon sok gyönyör őséget nyújt e -

Distribution of Woody Rosaceae in W. Asia XIII. Amygdalus Webbii Spach
ARBORETUM KÓRNICKIE Rocznik XIX — 1974 Kazimierz Browicz Distribution of Woody Rosaceae in W. Asia ХШ Amygdalus webbii Spach and closely related species The closely related group of almonds discussed below represents a group of species characterized by more or less thorny shoots (exception A. browiczii) and growth in the form of erect shrubs or small trees. They are frequently considered to be the original forms for A. communis, which numerous authors judge as being only a cultivated species. Howe ver this wiev is hard to accept in sipite of the fact that the true origin of A. communis is today difficult to determine. It seems however that it grows wild in southern Turkmenia in the Kopet-Dagh Mts., in major part of Turkey (particularity eastern), in western Iran, in western Syria, in Lebanon, in Israel and in western Jordan as well as possibly in southern Caucasus. It is striking that the ranges of the five species discussed here are separated from each other by frequently considerable distances yet they are all united through the range of A. communis. Thus at the wes tern extremity of A. communis range there falls the range of A. webbii, at the southern the range of A. korshinskyi, at the northern the range of A. fenzliana and at the eastern the range of A. haussknechtii and A. bro wiczii. 1. AMYGDALUS WEBBII SPACH Spach, Ann. Sci. Nat. Paris, 2 ser. 19 :117 (1843) Syn.: A. salicifolia Boiss. et Bal., in Boiss. Diagn. ser. 2 (6): 71 (1859). A. webbii Spach var. salicifolia (Boiss. et Bal.) Boiss., Fl. -

WO 2016/016826 Al 4 February 2016 (04.02.2016) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2016/016826 Al 4 February 2016 (04.02.2016) P O P C T (51) International Patent Classification: (74) Agent: xyAJ PARK; Level 22, State Insurance Tower, 1 A01H 1/06 (2006.01) C12N 15/61 (2006.01) Willis Street, Wellington (NZ). C12N 15/29 (2006.01) A01H 5/08 (2006.01) (81) Designated States (unless otherwise indicated, for every C12N 15/113 (2010.01) kind of national protection available): AE, AG, AL, AM, (21) International Application Number: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, PCT/IB2015/055743 BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (22) International Filing Date: HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, 30 July 2015 (30.07.2015) KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, (25) Filing Language: English MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, (26) Publication Language: English SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, (30) Priority Data: TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. 6282 14 1 August 2014 (01.08.2014) NZ (84) Designated States (unless otherwise indicated, for every (72) Inventors; and kind of regional protection available): ARIPO (BW, GH, (71) Applicants : DARE, Andrew Patrick [NZ/NZ]; 40 Jef GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, ferson Street, Glendowie, Auckland, 1071 (NZ). -
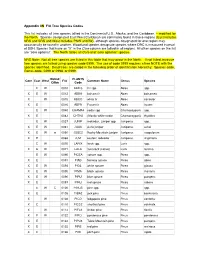
Appendix 3N FIA Tree Species Codes
Appendix 3N FIA Tree Species Codes This list includes all tree species tallied in the Continental U.S., Alaska, and the Caribbean. – modified for the North. Species designated East/West/Caribbean are commonly found in those regions (East includes NRS and SRS and West includes PNW and IW), although species designated for one region may occasionally be found in another. Woodland species designate species where DRC is measured instead of DBH. Species that have an “X” in the Core column are tallied in all regions. All other species on the list are “core optional”. The North tallies all Core and “core optional” species. NRS Note: Not all tree species are listed in this table that may occur in the North. If not listed, invasive tree species are tallied using species code 0999. The use of code 0999 requires a tree NOTE with the species identified. Dead trees are coded in the following order of identification hierarchy: Species code, Genus code, 0299 or 0998, or 0999. Wdlnd/ PLANTS Core East West FIA Common Name Genus Species Cribn Code E W 0010 ABIES Fir spp. Abies spp. X E W 0012 ABBA balsam fir Abies balsamea X W 0015 ABCO white fir Abies concolor X E 0016 ABFR Fraser fir Abies fraseri E W 0040 CHAMA4 cedar spp. Chamaecyparis spp. X E 0043 CHTH2 Atlantic white-cedar Chamaecyparis thyoides E W 0057 JUNIP redcedar, juniper spp. Juniperus spp. X E W 0061 JUAS Ashe juniper Juniperus ashei X E W w 0066 JUSC2 Rocky Mountain juniper Juniperus scopulorum X E 0068 JUVI eastern redcedar Juniperus virginiana E W 0070 LARIX larch spp.