Mouse Screen Reveals Multiple New Genes Underlying Mouse and Human Hearing Loss
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
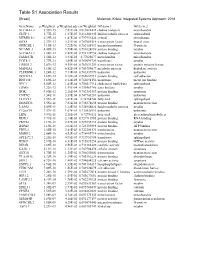
Table S1 Association Results
GeneName p.Weighted p.Weighted.adj cor.Weighted GO.term.1 GO.term.2 SLC44A1.2 3.55E-15 7.81E-08 0.819822422 choline transport mitochondria GLTP.1 5.77E-15 1.27E-07 0.816302445 lipid metabolic process sphingolipid MTMR10.1 6.39E-14 1.41E-06 0.797951424 cytosol phosphatase SOX8 2.37E-13 5.21E-06 0.787085514 transcription factor neural crest GPRC5B.1 4.19E-13 9.22E-06 0.782154937 integral membrane G-protein NCAM1.3 4.45E-13 9.79E-06 0.781624896 protein binding myelin SLC44A1.1 1.28E-12 2.82E-05 0.772132954 choline transport mitochondria FAM107B 1.56E-12 3.43E-05 0.77025677 mitochondria mitochondria UGT8.1 1.77E-12 3.89E-05 0.769099729 transferase myelin ERBB3.2 2.07E-12 4.55E-05 0.767631259 transcription factor protein tyrosine kinase MAN2A1 3.10E-12 6.82E-05 0.763759677 metabolic process hydrolase activity PLEKHH1.1 3.24E-12 7.13E-05 0.763337879 unknown unknown DOCK5.3 3.69E-12 8.12E-05 0.762087913 protein binding cell adhesion RNF130 3.69E-12 8.12E-05 0.762094156 membrane metal ion binding NPC1 6.50E-12 1.43E-04 0.756517114 cholesterol trafficking sphingolipid ERMN 7.22E-12 1.59E-04 0.755469786 actin binding myelin BOK 9.80E-12 2.16E-04 0.752383357 protein binding apoptosis CNTN2 1.54E-11 3.39E-04 0.747743281 unknown unknown ELOVL1 1.55E-11 3.41E-04 0.74764744 fatty acid sphingolipid DBNDD2 3.55E-11 7.81E-04 0.738878658 protein binding neuron projection LASS2 5.09E-11 1.12E-03 0.734954024 lipid metabolic process myelin C12orf34 7.57E-11 1.67E-03 0.730528911 unknown unknown LIPA 9.59E-11 2.11E-03 0.72786111 fatty acid glycerolipid metabolic -

SPNS2 Antibody (N-Term) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap17856a
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 SPNS2 Antibody (N-term) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP17856a Specification SPNS2 Antibody (N-term) - Product Information Application WB, IHC-P, FC,E Primary Accession Q8IVW8 Other Accession NP_001118230.1 Reactivity Human Host Rabbit Clonality Polyclonal Isotype Rabbit IgG Antigen Region 68-94 SPNS2 Antibody (N-term) - Additional Information Gene ID 124976 Other Names Protein spinster homolog 2, SPNS2 All lanes : Anti-SPNS2 Antibody (N-term) at Target/Specificity 1:1000 dilution Lane 1: human brain tissue This SPNS2 antibody is generated from lysate Lane 2: 293 whole cell lysate Lane 3: rabbits immunized with a KLH conjugated SK-BR-3 whole cell lysate Lysates/proteins at synthetic peptide between 68-94 amino 20 µg per lane. Secondary Goat Anti-Rabbit acids from the N-terminal region of human IgG, (H+L), Peroxidase conjugated at 1/10000 SPNS2. dilution. Predicted band size : 58 kDa Blocking/Dilution buffer: 5% NFDM/TBST. Dilution WB~~1:1000 IHC-P~~1:100 FC~~1:25 Format Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. Storage Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. Precautions SPNS2 Antibody (N-term) is for research use All lanes : Anti-SPNS2 Antibody (N-term) at only and not for use in diagnostic or 1:1000 dilution Lane 1: Human heart lysate therapeutic procedures. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

LRIG1 Gene Copy Number Analysis by Ddpcr and Correlations to Clinical
Faraz et al. BMC Cancer (2020) 20:459 https://doi.org/10.1186/s12885-020-06919-w RESEARCH ARTICLE Open Access LRIG1 gene copy number analysis by ddPCR and correlations to clinical factors in breast cancer Mahmood Faraz1, Andreas Tellström1, Christina Edwinsdotter Ardnor1, Kjell Grankvist2, Lukasz Huminiecki3,4, Björn Tavelin1, Roger Henriksson1, Håkan Hedman1 and Ingrid Ljuslinder1* Abstract Background: Leucine-rich repeats and immunoglobulin-like domains 1 (LRIG1) copy number alterations and unbalanced gene recombination events have been reported to occur in breast cancer. Importantly, LRIG1 loss was recently shown to predict early and late relapse in stage I-II breast cancer. Methods: We developed droplet digital PCR (ddPCR) assays for the determination of relative LRIG1 copy numbers and used these assays to analyze LRIG1 in twelve healthy individuals, 34 breast tumor samples previously analyzed by fluorescence in situ hybridization (FISH), and 423 breast tumor cytosols. Results: Four of the LRIG1/reference gene assays were found to be precise and robust, showing copy number ratios close to 1 (mean, 0.984; standard deviation, +/− 0.031) among the healthy control population. The correlation between the ddPCR assays and previous FISH results was low, possibly because of the different normalization strategies used. One in 34 breast tumors (2.9%) showed an unbalanced LRIG1 recombination event. LRIG1 copy number ratios were associated with the breast cancer subtype, steroid receptor status, ERBB2 status, tumor grade, and nodal status. Both LRIG1 loss and gain were associated with unfavorable metastasis-free survival; however, they did not remain significant prognostic factors after adjustment for common risk factors in the Cox regression analysis. -

LRIG1 Inhibits STAT3-Dependent Inflammation to Maintain Corneal Homeostasis
LRIG1 inhibits STAT3-dependent inflammation to maintain corneal homeostasis Takahiro Nakamura, … , Yann Barrandon, Shigeru Kinoshita J Clin Invest. 2014;124(1):385-397. https://doi.org/10.1172/JCI71488. Research Article Stem cells Corneal integrity and transparency are indispensable for good vision. Cornea homeostasis is entirely dependent upon corneal stem cells, which are required for complex wound-healing processes that restore corneal integrity following epithelial damage. Here, we found that leucine-rich repeats and immunoglobulin-like domains 1 (LRIG1) is highly expressed in the human holoclone-type corneal epithelial stem cell population and sporadically expressed in the basal cells of ocular-surface epithelium. In murine models, LRIG1 regulated corneal epithelial cell fate during wound repair. Deletion of Lrig1 resulted in impaired stem cell recruitment following injury and promoted a cell-fate switch from transparent epithelium to keratinized skin-like epidermis, which led to corneal blindness. In addition, we determined that LRIG1 is a negative regulator of the STAT3-dependent inflammatory pathway. Inhibition of STAT3 in corneas of Lrig1–/– mice rescued pathological phenotypes and prevented corneal opacity. Additionally, transgenic mice that expressed a constitutively active form of STAT3 in the corneal epithelium had abnormal features, including corneal plaques and neovascularization similar to that found in Lrig1–/– mice. Bone marrow chimera experiments indicated that LRIG1 also coordinates the function of bone marrow–derived inflammatory cells. Together, our data indicate that LRIG1 orchestrates corneal-tissue transparency and cell fate during repair, and identify LRIG1 as a key regulator of tissue homeostasis. Find the latest version: https://jci.me/71488/pdf Research article LRIG1 inhibits STAT3-dependent inflammation to maintain corneal homeostasis Takahiro Nakamura,1,2 Junji Hamuro,1 Mikiro Takaishi,3 Szandor Simmons,4 Kazuichi Maruyama,1 Andrea Zaffalon,5 Adam J. -

Anti-LRIG1 Antibody (ARG43047)
Product datasheet [email protected] ARG43047 Package: 50 μg anti-LRIG1 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes LRIG1 Tested Reactivity Hu Tested Application IHC-P, WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name LRIG1 Antigen Species Human Immunogen Synthetic peptide corresponding to a sequence of Human LRIG1. (AKRAFSGLESLEHLNLGENAIRSVQFDAFAKMKNLKELYI) Conjugation Un-conjugated Alternate Names LIG-1; LIG1; Leucine-rich repeats and immunoglobulin-like domains protein 1 Application Instructions Application table Application Dilution IHC-P 1:200 - 1:1000 WB 1:500 - 1:2000 Application Note IHC-P: Antigen Retrieval: Heat mediation was performed in Citrate buffer (pH 6.0) for 20 min. * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Calculated Mw 119 kDa Properties Form Liquid Purification Affinity purification with immunogen. Buffer 0.2% Na2HPO4, 0.9% NaCl, 0.05% Sodium azide and 4% Trehalose. Preservative 0.05% Sodium azide Stabilizer 4% Trehalose Concentration 0.5 - 1 mg/ml Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C or below. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. www.arigobio.com 1/2 Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Gene Symbol LRIG1 Gene Full Name leucine-rich repeats and immunoglobulin-like domains 1 Function Acts as a feedback negative regulator of signaling by receptor tyrosine kinases, through a mechanism that involves enhancement of receptor ubiquitination and accelerated intracellular degradation. -

Function in Immune System Spns2 Transporter the Role of Sphingosine-1-Phosphate
The Journal of Immunology The Role of Sphingosine-1-Phosphate Transporter Spns2 in Immune System Function Anastasia Nijnik,*,† Simon Clare,* Christine Hale,* Jing Chen,* Claire Raisen,* Lynda Mottram,* Mark Lucas,* Jeanne Estabel,* Edward Ryder,* Hibret Adissu,‡ Sanger Mouse Genetics Project,1 Niels C. Adams,* Ramiro Ramirez-Solis,* Jacqueline K. White,* Karen P. Steel,* Gordon Dougan,* and Robert E. W. Hancock† Sphingosine-1-phosphate (S1P) is lipid messenger involved in the regulation of embryonic development, immune system functions, and many other physiological processes. However, the mechanisms of S1P transport across cellular membranes remain poorly understood, with several ATP-binding cassette family members and the spinster 2 (Spns2) member of the major facilitator superfamily known to mediate S1P transport in cell culture. Spns2 was also shown to control S1P activities in zebrafish in vivo and to play a critical role in zebrafish cardiovascular development. However, the in vivo roles of Spns2 in mammals and its involvement in the different S1P-dependent physiological processes have not been investigated. In this study, we charac- terized Spns2-null mouse line carrying the Spns2tm1a(KOMP)Wtsi allele (Spns2tm1a). The Spns2tm1a/tm1a animals were viable, indi- cating a divergence in Spns2 function from its zebrafish ortholog. However, the immunological phenotype of the Spns2tm1a/tm1a mice closely mimicked the phenotypes of partial S1P deficiency and impaired S1P-dependent lymphocyte trafficking, with a depletion of lymphocytes in circulation, an increase in mature single-positive T cells in the thymus, and a selective reduction in mature B cells in the spleen and bone marrow. Spns2 activity in the nonhematopoietic cells was critical for normal lymphocyte development and localization. -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -

Multiple Pathways for Protein Transport to Peroxisomes
Review Multiple Pathways for Protein Transport to Peroxisomes P.K. Kim 1,2 and E.H. Hettema 3 1 - Program in Cell Biology, Hospital for Sick Children, Toronto, ON, Canada M5G 1X8 2 - Department of Biochemistry, University of Toronto, Toronto, ON, Canada M5S 1A8 3 - Department of Molecular Biology and Biotechnology, University of Sheffield, Firth Court, Western Bank, Sheffield, South Yorkshire S10 2TN, United Kingdom Correspondence to E.H. Hettema: [email protected] http://dx.doi.org/10.1016/j.jmb.2015.02.005 Edited by S. High Abstract Peroxisomes are unique among the organelles of the endomembrane system. Unlike other organelles that derive most if not all of their proteins from the ER (endoplasmic reticulum), peroxisomes contain dedicated machineries for import of matrix proteins and insertion of membrane proteins. However, peroxisomes are also able to import a subset of their membrane proteins from the ER. One aspect of peroxisome biology that has remained ill defined is the role the various import pathways play in peroxisome maintenance. In this review, we discuss the available data on matrix and membrane protein import into peroxisomes. © 2015 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/). Introduction dependent on consumption of energy in the form of ATP. Several aspects of peroxisomal protein transport Peroxisomes are organelles found in almost all distinguish it from other translocation systems. For eukaryotic cells. They are bounded by a single example, peroxisomal proteins can fold, acquire membrane and are usually spherical. -

High Constitutive Cytokine Release by Primary Human Acute Myeloid Leukemia Cells Is Associated with a Specific Intercellular Communication Phenotype
Supplementary Information High Constitutive Cytokine Release by Primary Human Acute Myeloid Leukemia Cells Is Associated with a Specific Intercellular Communication Phenotype Håkon Reikvam 1,2,*, Elise Aasebø 1, Annette K. Brenner 2, Sushma Bartaula-Brevik 1, Ida Sofie Grønningsæter 2, Rakel Brendsdal Forthun 2, Randi Hovland 3,4 and Øystein Bruserud 1,2 1 Department of Clinical Science, University of Bergen, 5020, Bergen, Norway 2 Department of Medicine, Haukeland University Hospital, 5021, Bergen, Norway 3 Department of Medical Genetics, Haukeland University Hospital, 5021, Bergen, Norway 4 Institute of Biomedicine, University of Bergen, 5020, Bergen, Norway * Correspondence: [email protected]; Tel.: +55-97-50-00 J. Clin. Med. 2019, 8, x 2 of 36 Figure S1. Mutational studies in a cohort of 71 AML patients. The figure shows the number of patients with the various mutations (upper), the number of mutations in for each patient (middle) and the number of main classes with mutation(s) in each patient (lower). 2 J. Clin. Med. 2019, 8, x; doi: www.mdpi.com/journal/jcm J. Clin. Med. 2019, 8, x 3 of 36 Figure S2. The immunophenotype of primary human AML cells derived from 62 unselected patients. The expression of the eight differentiation markers CD13, CD14, CD15, CD33, CD34, CD45, CD117 and HLA-DR was investigated for 62 of the 71 patients included in our present study. We performed an unsupervised hierarchical cluster analysis and identified four patient main clusters/patient subsets. The mutational profile for each f the 62 patients is also given (middle), no individual mutation of main class of mutations showed any significant association with any of the for differentiation marker clusters (middle). -

Network Pharmacology Interpretation of Fuzheng–Jiedu Decoction Against Colorectal Cancer
Hindawi Evidence-Based Complementary and Alternative Medicine Volume 2021, Article ID 4652492, 16 pages https://doi.org/10.1155/2021/4652492 Research Article Network Pharmacology Interpretation of Fuzheng–Jiedu Decoction against Colorectal Cancer Hongshuo Shi ,1 Sisheng Tian,2 and Hu Tian 3 1College of Traditional Chinese Medicine, Shandong University of Traditional Chinese Medicine, Jinan, Shandong, China 2School of Management, Shandong University of Traditional Chinese Medicine, Jinan, Shandong, China 3College of Traditional Chinese Medicine, Shandong University of Traditional Chinese Medicine, Jinan, Shandong, China Correspondence should be addressed to Hu Tian; [email protected] Received 7 April 2020; Revised 3 January 2021; Accepted 21 January 2021; Published 20 February 2021 Academic Editor: George B. Lenon Copyright © 2021 Hongshuo Shi et al. ,is is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Introduction. Traditional Chinese medicine (TCM) believes that the pathogenic factors of colorectal cancer (CRC) are “deficiency, dampness, stasis, and toxin,” and Fuzheng–Jiedu Decoction (FJD) can resist these factors. In this study, we want to find out the potential targets and pathways of FJD in the treatment of CRC and also explain from a scientific point of view that FJD multidrug combination can resist “deficiency, dampness, stasis, and toxin.” Methods. We get the composition of FJD from the TCMSP database and get its potential target. We also get the potential target of colorectal cancer according to the OMIM Database, TTD Database, GeneCards Database, CTD Database, DrugBank Database, and DisGeNET Database. -

Characterization of the Macrophage Transcriptome in Glomerulonephritis-Susceptible and -Resistant Rat Strains
Genes and Immunity (2011) 12, 78–89 & 2011 Macmillan Publishers Limited All rights reserved 1466-4879/11 www.nature.com/gene ORIGINAL ARTICLE Characterization of the macrophage transcriptome in glomerulonephritis-susceptible and -resistant rat strains K Maratou1, J Behmoaras2, C Fewings1, P Srivastava1, Z D’Souza1, J Smith3, L Game4, T Cook2 and T Aitman1 1Physiological Genomics and Medicine Group, MRC Clinical Sciences Centre, Imperial College London, London, UK; 2Centre for Complement and Inflammation Research, Imperial College London, London, UK; 3Renal Section, Imperial College London, London, UK and 4Genomics Laboratory, MRC Clinical Sciences Centre, London, UK Crescentic glomerulonephritis (CRGN) is a major cause of rapidly progressive renal failure for which the underlying genetic basis is unknown. Wistar–Kyoto (WKY) rats show marked susceptibility to CRGN, whereas Lewis rats are resistant. Glomerular injury and crescent formation are macrophage dependent and mainly explained by seven quantitative trait loci (Crgn1–7). Here, we used microarray analysis in basal and lipopolysaccharide (LPS)-stimulated macrophages to identify genes that reside on pathways predisposing WKY rats to CRGN. We detected 97 novel positional candidates for the uncharacterized Crgn3–7. We identified 10 additional secondary effector genes with profound differences in expression between the two strains (45-fold change, o1% false discovery rate) for basal and LPS-stimulated macrophages. Moreover, we identified eight genes with differentially expressed alternatively spliced isoforms, by using an in-depth analysis at the probe level that allowed us to discard false positives owing to polymorphisms between the two rat strains. Pathway analysis identified several common linked pathways, enriched for differentially expressed genes, which affect macrophage activation.