Recommendations for Anti-Microbial Use for Secondary Infections in Patients with COVID-19 Rapid Review
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Therapeutic Alternatives for Drug-Resistant Cabapenemase- Producing Enterobacteria
THERAPEUTIC ALTERNATIVES FOR MULTIDRUG-RESISTANT AND EXTREMELY All isolates resistant to carbapenems were evaluated Fig. 1: Susceptibility (%) of Carbapenemase-producing MDRE Fig. 3: Susceptibility (%) of VIM-producing MDRE to Alternative DRUG-RESISTANT CABAPENEMASE- for the presence of genes encoding carbapenemases to Alternative Antibiotics Antibiotics PRODUCING ENTEROBACTERIA OXA-48-like, KPC, GES, NDM, VIM, IMP and GIM. M. Almagro*, A. Kramer, B. Gross, S. Suerbaum, S. Schubert. Max Von Pettenkofer Institut, Faculty of Medicine. LMU Munich, München, Germany. RESULTS BACKROUND 44 isolates of CPE were collected: OXA-48 (n=29), VIM (n=9), NDM-1 (n=5), KPC (n=1) and GES (n=1). The increasing emergence and dissemination of carbapenem-resistant gram-negative bacilli has From the 44 CPE isolates, 26 isolates were identified reduced significantly the options for sufficient as Klebsiella pneumoniae (68% of the OXA-48 CPE), 8 Fig. 2: Susceptibility (%) of OXA-48-producing MDRE to Fig. 4: Susceptibility (%) of NDM-producing MDRE to Alternative antibiotic therapy. The genes encoding most of these as Escherichia coli, 6 as Enterobacter cloacae, 2 as Alternative Antibiotics Antibiotics carbapenemases reside on plasmids or transposons Citrobacter freundii,1 as Providencia stuartii and 1 as carrying additional resistance genes which confer Morganella morganii. multidrug resistance to the isolates. 31 isolates (70%) were causing an infection, including urinary tract infection (20%), respiratory tract MATERIALS AND METHODS infection (18%), abdominal infection (18%), bacteraemia (9%) and skin and soft tissue infection In the present study, we tested the in vitro activity of (5%). 13 isolates were believed to be colonizers. antimicrobial agents against a well-characterized c o l l e c t i o n o f c a r b a p e n e m a s e - p r o d u c i n g Isolates were classified as 32 MDRE and 13 XDRE. -

FDA-Approved Drugs with Potent in Vitro Antiviral Activity Against Severe Acute Respiratory Syndrome Coronavirus 2
pharmaceuticals Article FDA-Approved Drugs with Potent In Vitro Antiviral Activity against Severe Acute Respiratory Syndrome Coronavirus 2 1, , 1, 2 1 Ahmed Mostafa * y , Ahmed Kandeil y , Yaseen A. M. M. Elshaier , Omnia Kutkat , Yassmin Moatasim 1, Adel A. Rashad 3 , Mahmoud Shehata 1 , Mokhtar R. Gomaa 1, Noura Mahrous 1, Sara H. Mahmoud 1, Mohamed GabAllah 1, Hisham Abbas 4 , Ahmed El Taweel 1, Ahmed E. Kayed 1, Mina Nabil Kamel 1, Mohamed El Sayes 1, Dina B. Mahmoud 5 , Rabeh El-Shesheny 1 , Ghazi Kayali 6,7,* and Mohamed A. Ali 1,* 1 Center of Scientific Excellence for Influenza Viruses, National Research Centre, Giza 12622, Egypt; [email protected] (A.K.); [email protected] (O.K.); [email protected] (Y.M.); [email protected] (M.S.); [email protected] (M.R.G.); [email protected] (N.M.); [email protected] (S.H.M.); [email protected] (M.G.); [email protected] (A.E.T.); [email protected] (A.E.K.); [email protected] (M.N.K.); [email protected] (M.E.S.); [email protected] (R.E.-S.) 2 Organic & Medicinal Chemistry Department, Faculty of Pharmacy, University of Sadat City, Menoufia 32897, Egypt; [email protected] 3 Department of Biochemistry & Molecular Biology, Drexel University College of Medicine, Philadelphia, PA 19102, USA; [email protected] 4 Department of Microbiology and Immunology, Zagazig University, Zagazig 44519, Egypt; [email protected] 5 Pharmaceutics Department, National Organization for Drug Control and Research, Giza 12654, Egypt; [email protected] 6 Department of Epidemiology, Human Genetics, and Environmental Sciences, University of Texas, Houston, TX 77030, USA 7 Human Link, Baabda 1109, Lebanon * Correspondence: [email protected] (A.M.); [email protected] (G.K.); [email protected] (M.A.A.) Contributed equally to this work. -

University of Cincinnati
UNIVERSITY OF CINCINNATI Date: February 22, 2007 I, _ Samuel Lee Hayes________________________________________, hereby submit this work as part of the requirements for the degree of: Doctor of Philosophy in: Biological Sciences It is entitled: Response of Mammalian Models to Exposure of Bacteria from the Genus Aeromonas Evaluated using Transcriptional Analysis and Conjectures on Disease Mechanisms This work and its defense approved by: Chair: _Brian K. Kinkle _Dennis W. Grogan _Richard D. Karp _Mario Medvedovic _Stephen J. Vesper Response of Mammalian Models to Exposure of Bacteria from the Genus Aeromonas Evaluated using Transcriptional Analysis and Conjectures on Disease Mechanisms A dissertation submitted to the Division of Graduate Studies and Research of the University of Cincinnati in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in the Department of Biological Sciences of the College of Arts and Sciences 2007 by Samuel Lee Hayes B.S. Ohio University, 1978 M.S. University of Cincinnati, 1986 Committee Chair: Dr. Brian K. Kinkle Abstract The genus Aeromonas contains virulent bacteria implicated in waterborne disease, as well as avirulent strains. One of my research objectives was to identify and characterize host- pathogen relationships specific to Aeromonas spp. Aeromonas virulence was assessed using changes in host mRNA expression after infecting cell cultures and live animals. Messenger RNA extracts were hybridized to murine genomic microarrays. Initially, these model systems were infected with two virulent A. hydrophila strains, causing up-regulation of over 200 and 50 genes in animal and cell culture tissues, respectively. Twenty-six genes were common between the two model systems. The live animal model was used to define virulence for many Aeromonas spp. -

Genetic Diversity of Bartonella Species in Small Mammals in the Qaidam
www.nature.com/scientificreports OPEN Genetic diversity of Bartonella species in small mammals in the Qaidam Basin, western China Huaxiang Rao1, Shoujiang Li3, Liang Lu4, Rong Wang3, Xiuping Song4, Kai Sun5, Yan Shi3, Dongmei Li4* & Juan Yu2* Investigation of the prevalence and diversity of Bartonella infections in small mammals in the Qaidam Basin, western China, could provide a scientifc basis for the control and prevention of Bartonella infections in humans. Accordingly, in this study, small mammals were captured using snap traps in Wulan County and Ge’ermu City, Qaidam Basin, China. Spleen and brain tissues were collected and cultured to isolate Bartonella strains. The suspected positive colonies were detected with polymerase chain reaction amplifcation and sequencing of gltA, ftsZ, RNA polymerase beta subunit (rpoB) and ribC genes. Among 101 small mammals, 39 were positive for Bartonella, with the infection rate of 38.61%. The infection rate in diferent tissues (spleens and brains) (χ2 = 0.112, P = 0.738) and gender (χ2 = 1.927, P = 0.165) of small mammals did not have statistical diference, but that in diferent habitats had statistical diference (χ2 = 10.361, P = 0.016). Through genetic evolution analysis, 40 Bartonella strains were identifed (two diferent Bartonella species were detected in one small mammal), including B. grahamii (30), B. jaculi (3), B. krasnovii (3) and Candidatus B. gerbillinarum (4), which showed rodent-specifc characteristics. B. grahamii was the dominant epidemic strain (accounted for 75.0%). Furthermore, phylogenetic analysis showed that B. grahamii in the Qaidam Basin, might be close to the strains isolated from Japan and China. -

Novel Small Rnas Expressed by Bartonella Bacilliformis Under Multiple Conditions 2 Reveal Potential Mechanisms for Persistence in the Sand Fly Vector and Human 3 Host
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.04.235903; this version posted August 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Novel small RNAs expressed by Bartonella bacilliformis under multiple conditions 2 reveal potential mechanisms for persistence in the sand fly vector and human 3 host 4 5 6 Shaun Wachter1, Linda D. Hicks1, Rahul Raghavan2 and Michael F. Minnick1* 7 8 9 10 1 Program in Cellular, Molecular & Microbial Biology, Division of Biological Sciences, 11 University of Montana, Missoula, Montana, United States of America 12 2 Department of Biology and Center for Life in Extreme Environments, Portland State 13 University, Portland, Oregon, United States of America 14 15 16 17 * Corresponding author 18 E-mail: [email protected] (MFM) 19 20 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.04.235903; this version posted August 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 21 Abstract 22 Bartonella bacilliformis, the etiological agent of Carrión’s disease, is a Gram-negative, 23 facultative intracellular alphaproteobacterium. Carrión’s disease is an emerging but neglected 24 tropical illness endemic to Peru, Colombia, and Ecuador. B. bacilliformis is spread between 25 humans through the bite of female phlebotomine sand flies. -

09 Piqueras.Qxp
PERSPECTIVES INTERNATIONAL MICROBIOLOGY (2007) 10:217-226 DOI: 10.2436/20.1501.01.30 ISSN: 1139-6709 www.im.microbios.org Microbiology: a dangerous profession? Mercè Piqueras President, Catalan Association for Science Communication (ACCC), Barcelona, Spain The history of science contains many cases of researchers eases in Minorca from the year 1744 to 1749 to which is pre- who have died because of their professional activity. In the fixed, a short account of the climate, productions, inhabi- field of microbiology, some have died or have come close to tants, and endemical distempers of that island (T. Cadell, D. death from infection by agents that were the subject of their Wilson and G. Nicol, London, 1751), which he dedicated to research (Table 1). Infections that had a lethal outcome were the Society of Surgeons of His Majesty’s Royal Navy. usually accidental. Sometimes, however, researchers inocu- Minorcan historian of science Josep M. Vidal Hernández has lated themselves with the pathogen or did not take preventive described and carefully analyzed Cleghorn’s work in measures against the potential pathogen because they wanted Minorca and his report [43]. According to Vidal, what to prove their hypotheses—or disprove someone else’s— Cleghorn describes is “tertian” fever, which was the name regarding the origin of the infection. Here is an overview of given at the time to fever caused by malaria parasites with a several episodes in the history of microbiology since the mid periodicity of 48 hours. In fact, Cleghorn used quinine to nineteenth century involving researchers or workers in fields treat tertian fever (i.e, malaria), which was not eradicated related to microbiology who have become infected. -

Severe Sepsis and Septic Shock Antibiotic Guide
Stanford Health Issue Date: 05/2017 Stanford Antimicrobial Safety and Sustainability Program Severe Sepsis and Septic Shock Antibiotic Guide Table 1: Antibiotic selection options for healthcare associated and/or immunocompromised patients • Healthcare associated: intravenous therapy, wound care, or intravenous chemotherapy within the prior 30 days, residence in a nursing home or other long-term care facility, hospitalization in an acute care hospital for two or more days within the prior 90 days, attendance at a hospital or hemodialysis clinic within the prior 30 days • Immunocompromised: Receiving chemotherapy, known systemic cancer not in remission, ANC <500, severe cell-mediated immune deficiency Table 2: Antibiotic selection options for community acquired, immunocompetent patients Table 3: Antibiotic selection options for patients with simple sepsis, community acquired, immunocompetent patients requiring hospitalization. Risk Factors for Select Organisms P. aeruginosa MRSA Invasive Candidiasis VRE (and other resistant GNR) Community acquired: • Known colonization with MDROs • Central venous catheter • Liver transplant • Prior IV antibiotics within 90 day • Recent MRSA infection • Broad-spectrum antibiotics • Known colonization • Known colonization with MDROs • Known MRSA colonization • + 1 of the following risk factors: • Prolonged broad antibacterial • Skin & Skin Structure and/or IV access site: ♦ Parenteral nutrition therapy Hospital acquired: ♦ Purulence ♦ Dialysis • Prolonged profound • Prior IV antibiotics within 90 days ♦ Abscess -

Mechanisms of Resistance to P-Lactam Antibiotics Amongst 1993
J. Med. Microbiol. - Vol. 43 (1999, 30G309 0 1995 The Pathological Society of Great Britain and Ireland ANTI MICROBIAL AGENTS Mechanisms of resistance to p-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993 H. Y. CHEN, ME1 YUAN and D. M. LIVERMORE Department of Medical Microbiology, The London Hospital Medical College, Turner Street, London El 2AD Summary. Antimicrobial resistance among 199 1 Pseudomonas aeruginosa isolates collected at 24 UK hospitals during late 1993 was surveyed. Three-hundred and seventy-two of the isolates were resistant, or had reduced susceptibility, to some or all of azlocillin, carbenicillin, ceftazidime, imipenem and meropenem, and the mechanisms underlying their behaviour were examined. Only 13 isolates produced secondary p-lactamases : six possessed PSE- 1 or PSE-4 enzymes and seven had novel OXA enzyme types. Those with PSE types were highly resistant to azlocillin and carbenicillin whereas those with OXA enzymes were less resistant to these penicillins. Chromosomal p-lactamase derepression was demonstrated in 54 isolates, most of which were resistant to ceftazidime and azlocillin although susceptible to carbenicillin and carbapenems. p-Lactamase-independent “ intrinsic” resistance occurred in 277 isolates and is believed to reflect some combination of impermeability and efflux. Two forms were seen : the classical type, present in 195 isolates, gave carbenicillin resistance (MIC > 128 mg/L) and reduced susceptibility to ciprofloxacin and to all p-lactam agents except imipenem; a novel variant, seen in 82 isolates, affected only azlocillin, ceftazidime and, to a small extent, meropenem. Resistance to imipenem was largely dissociated from that to other p-lactam agents, and probably reflected loss of D2 porin, whereas resistance to meropenem was mostly associated with intrinsic resistance to penicillins and cephalosporins. -

Antibiotic Assay Medium No. 3 (Assay Broth) Is Used for Microbiological Assay of Antibiotics. M042
HiMedia Laboratories Technical Data Antibiotic Assay Medium No. 3 (Assay Broth) is used for M042 microbiological assay of antibiotics. Antibiotic Assay Medium No. 3 (Assay Broth) is used for microbiological assay of antibiotics. Composition** Ingredients Gms / Litre Peptic digest of animal tissue (Peptone) 5.000 Beef extract 1.500 Yeast extract 1.500 Dextrose 1.000 Sodium chloride 3.500 Dipotassium phosphate 3.680 Potassium dihydrogen phosphate 1.320 Final pH ( at 25°C) 7.0±0.2 **Formula adjusted, standardized to suit performance parameters Directions Suspend 17.5 grams in 1000 ml distilled water. Heat if necessary to dissolve the medium completely. Sterilize by autoclaving at 15 lbs pressure (121°C) for 15 minutes. Advice:Recommended for the Microbiological assay of Amikacin, Bacitracin, Capreomycin, Chlortetracycline,Chloramphenicol,Cycloserine,Demeclocycline,Dihydrostreptomycin, Doxycycline, Gentamicin, Gramicidin, Kanamycin, Methacycline, Neomycin, Novobiocin, Oxytetracycline, Rolitetracycline, Streptomycin, Tetracycline, Tobramycin, Trolendomycin and Tylosin according to official methods . Principle And Interpretation Antibiotic Assay Medium is used in the performance of antibiotic assays. Grove and Randall have elucidated those antibiotic assays and media in their comprehensive treatise on antibiotic assays (1). Antibiotic Assay Medium No. 3 (Assay Broth) is used in the microbiological assay of different antibiotics in pharmaceutical and food products by the turbidimetric method. Ripperre et al reported that turbidimetric methods for determining the potency of antibiotics are inherently more accurate and more precise than agar diffusion procedures (2). Turbidimetric antibiotic assay is based on the change or inhibition of growth of a test microorganims in a liquid medium containing a uniform concentration of an antibiotic. After incubation of the test organism in the working dilutions of the antibiotics, the amount of growth is determined by measuring the light transmittance using spectrophotometer. -
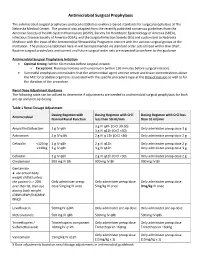
Antimicrobial Surgical Prophylaxis
Antimicrobial Surgical Prophylaxis The antimicrobial surgical prophylaxis protocol establishes evidence-based standards for surgical prophylaxis at The Nebraska Medical Center. The protocol was adapted from the recently published consensus guidelines from the American Society of Health-System Pharmacists (ASHP), Society for Healthcare Epidemiology of America (SHEA), Infectious Disease Society of America (IDSA), and the Surgical Infection Society (SIS) and customized to Nebraska Medicine with the input of the Antimicrobial Stewardship Program in concert with the various surgical groups at the institution. The protocol established here-in will be implemented via standard order sets utilized within One Chart. Routine surgical prophylaxis and current and future surgical order sets are expected to conform to this guidance. Antimicrobial Surgical Prophylaxis Initiation Optimal timing: Within 60 minutes before surgical incision o Exceptions: Fluoroquinolones and vancomycin (within 120 minutes before surgical incision) Successful prophylaxis necessitates that the antimicrobial agent achieve serum and tissue concentrations above the MIC for probable organisms associated with the specific procedure type at the time of incision as well as for the duration of the procedure. Renal Dose Adjustment Guidance The following table can be utilized to determine if adjustments are needed to antimicrobial surgical prophylaxis for both pre-op and post-op dosing. Table 1 Renal Dosage Adjustment Dosing Regimen with Dosing Regimen with CrCl Dosing Regimen with -

Bartonella Henselae • Fleas and Black-Legged Ticks (Also Called Deer Ticks) of the Genus Ixodes May Serve As Vectors, but This Has Not Disease Agent: Been Proven
APPENDIX 2 Bartonella henselae • Fleas and black-legged ticks (also called deer ticks) of the genus Ixodes may serve as vectors, but this has not Disease Agent: been proven. • Bartonella henselae Blood Phase: Disease Agent Characteristics: • Agent found in endothelial cells and associated with RBCs in symptomatic cases • Gram-negative bacillus or coccobacillus, aerobic, • Occult bacteremia sometimes occurs in the absence nonmotile, nonspore-forming, facultatively intracel- of specific antibodies. lular bacterium • Order: Rhizobiales; Family: Bartonellaceae Survival/Persistence in Blood Products: • Size: 0.3-0.6 ¥ 0.3-1.0 mm • Nucleic acid: Approximately 1900 kb of DNA • A spiking study suggests that B. henselae added to RBCs can be recovered on solid media through 35 Disease Name: days of storage at 4°C. • Cat scratch disease • Cat scratch fever Transmission by Blood Transfusion: • Bacillary angiomatosis • Theoretical • Bacillary peliosis Cases/Frequency in Population: Priority Level: • 22,000 cases per year estimated in the US • Scientific/Epidemiologic evidence regarding blood • 2-6% in US blood donors safety: Theoretical • Cumulative seroprevalence of 7.1% to B. henselae and • Public perception and/or regulatory concern regard- B. quintana in US veterinary professionals ing blood safety: Absent • Public concern regarding disease agent: Very low Incubation Period: Background: • 3-10 days to appearance of papule at inoculation site; regional adenopathy may follow after a few weeks • In 1909,ALBartondescribed organisms that adhered to RBCs. Likelihood of Clinical Disease: • The name Bartonella bacilliformis was used for the • Relatively benign and self-limiting, lasting 6-12 weeks only member of the group identified before 1993. in the absence of antibiotic therapy • Several other species of Bartonella are known to infect humans, but at present, B. -

Delineation of Aeromonas Hydrophila Pathotypes by Dectection of Putative Virulence Factors Using Polymerase Chain Reaction and N
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by DigitalCommons@Kennesaw State University Kennesaw State University DigitalCommons@Kennesaw State University Master of Science in Integrative Biology Theses Biology & Physics Summer 7-20-2015 Delineation of Aeromonas hydrophila Pathotypes by Dectection of Putative Virulence Factors using Polymerase Chain Reaction and Nematode Challenge Assay John Metz Kennesaw State University, [email protected] Follow this and additional works at: http://digitalcommons.kennesaw.edu/integrbiol_etd Part of the Integrative Biology Commons Recommended Citation Metz, John, "Delineation of Aeromonas hydrophila Pathotypes by Dectection of Putative Virulence Factors using Polymerase Chain Reaction and Nematode Challenge Assay" (2015). Master of Science in Integrative Biology Theses. Paper 7. This Thesis is brought to you for free and open access by the Biology & Physics at DigitalCommons@Kennesaw State University. It has been accepted for inclusion in Master of Science in Integrative Biology Theses by an authorized administrator of DigitalCommons@Kennesaw State University. For more information, please contact [email protected]. Delineation of Aeromonas hydrophila Pathotypes by Detection of Putative Virulence Factors using Polymerase Chain Reaction and Nematode Challenge Assay John Michael Metz Submitted in partial fulfillment of the requirements for the Master of Science Degree in Integrative Biology Thesis Advisor: Donald J. McGarey, Ph.D Department of Molecular and Cellular Biology Kennesaw State University ABSTRACT Aeromonas hydrophila is a Gram-negative, bacterial pathogen of humans and other vertebrates. Human diseases caused by A. hydrophila range from mild gastroenteritis to soft tissue infections including cellulitis and acute necrotizing fasciitis. When seen in fish it causes dermal ulcers and fatal septicemia, which are detrimental to aquaculture stocks and has major economic impact to the industry.