The 23S Ribosomal RNA from Pyrococcus Furiosus Is Circularly
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Domain of Escherichia Coli 16S Ribosomal RNA Using Site-Directed Photoaffinity Crosslinking
Downloaded from rnajournal.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press RNA (1998), 4:1455–1466+ Cambridge University Press+ Printed in the USA+ Copyright © 1998 RNA Society+ Analysis of the conformation of the 39 major domain of Escherichia coli 16S ribosomal RNA using site-directed photoaffinity crosslinking ALEXANDRE MONTPETIT,1 CATHERINE PAYANT,1 JAMES M. NOLAN,2 and LÉA BRAKIER-GINGRAS1 1Département de Biochimie, Université de Montréal, Montréal, Québec H3T 1J4, Canada 2Department of Biochemistry, Tulane University Medical Center, New Orleans, Louisiana 70112, USA ABSTRACT The 39 major domain of Escherichia coli 16S rRNA, which occupies the head of the small ribosomal subunit, is involved in several functions of the ribosome. We have used a site-specific crosslinking procedure to gain further insights into the higher-order structure of this domain. Circularly permuted RNAs were used to introduce an azi- dophenacyl group at specific positions within the 39 major domain. Crosslinks were generated in a high-ionic strength buffer that has been used for ribosome reconstitution studies and so enables the RNA to adopt a structure recognized by ribosomal proteins. The crosslinking sites were identified by primer extension and confirmed by assessing the mobility of the crosslinked RNA lariats in denaturing polyacrylamide gels. Eight crosslinks were characterized. Among them, one crosslink demonstrates that helix 28 is proximal to the top of helix 34, and two others show that the 1337 region, located in an internal loop at the junction of helices 29, 30, 41, and 42, is proximal to the center of helix 30 and to a segment connecting helix 28 to helix 29. -

Crystal Structure of the Eukaryotic 60S Ribosomal Subunit in Complex with Initiation Factor 6
Research Collection Doctoral Thesis Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6 Author(s): Voigts-Hoffmann, Felix Publication Date: 2012 Permanent Link: https://doi.org/10.3929/ethz-a-007303759 Rights / License: In Copyright - Non-Commercial Use Permitted This page was generated automatically upon download from the ETH Zurich Research Collection. For more information please consult the Terms of use. ETH Library ETH Zurich Dissertation No. 20189 Crystal Structure of the Eukaryotic 60S Ribosomal Subunit in Complex with Initiation Factor 6 A dissertation submitted to ETH ZÜRICH for the degree of Doctor of Sciences (Dr. sc. ETH Zurich) presented by Felix Voigts-Hoffmann MSc Molecular Biotechnology, Universität Heidelberg born April 11, 1981 citizen of Göttingen, Germany accepted on recommendation of Prof. Dr. Nenad Ban (Examiner) Prof. Dr. Raimund Dutzler (Co-examiner) Prof. Dr. Rudolf Glockshuber (Co-examiner) 2012 blank page ii Summary Ribosomes are large complexes of several ribosomal RNAs and dozens of proteins, which catalyze the synthesis of proteins according to the sequence encoded in messenger RNA. Over the last decade, prokaryotic ribosome structures have provided the basis for a mechanistic understanding of protein synthesis. While the core functional centers are conserved in all kingdoms, eukaryotic ribosomes are much larger than archaeal or bacterial ribosomes. Eukaryotic ribosomal rRNA and proteins contain extensions or insertions to the prokaryotic core, and many eukaryotic proteins do not have prokaryotic counterparts. Furthermore, translation regulation and ribosome biogenesis is much more complex in eukaryotes, and defects in components of the translation machinery are associated with human diseases and cancer. -

Micrornas Mediated Regulation of the Ribosomal Proteins and Its Consequences on the Global Translation of Proteins
cells Review microRNAs Mediated Regulation of the Ribosomal Proteins and Its Consequences on the Global Translation of Proteins Abu Musa Md Talimur Reza 1,2 and Yu-Guo Yuan 1,3,* 1 Jiangsu Co-Innovation Center of Prevention and Control of Important Animal Infectious Diseases and Zoonoses, College of Veterinary Medicine, Yangzhou University, Yangzhou 225009, China; [email protected] 2 Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawi´nskiego5a, 02-106 Warsaw, Poland 3 Jiangsu Key Laboratory of Zoonosis/Joint International Research Laboratory of Agriculture and Agri-Product Safety, The Ministry of Education of China, Yangzhou University, Yangzhou 225009, China * Correspondence: [email protected]; Tel.: +86-514-8797-9228 Abstract: Ribosomal proteins (RPs) are mostly derived from the energy-consuming enzyme families such as ATP-dependent RNA helicases, AAA-ATPases, GTPases and kinases, and are important structural components of the ribosome, which is a supramolecular ribonucleoprotein complex, composed of Ribosomal RNA (rRNA) and RPs, coordinates the translation and synthesis of proteins with the help of transfer RNA (tRNA) and other factors. Not all RPs are indispensable; in other words, the ribosome could be functional and could continue the translation of proteins instead of lacking in some of the RPs. However, the lack of many RPs could result in severe defects in the biogenesis of ribosomes, which could directly influence the overall translation processes and global expression of the proteins leading to the emergence of different diseases including cancer. While microRNAs (miRNAs) are small non-coding RNAs and one of the potent regulators of the post-transcriptional 0 gene expression, miRNAs regulate gene expression by targeting the 3 untranslated region and/or coding region of the messenger RNAs (mRNAs), and by interacting with the 50 untranslated region, Citation: Reza, A.M.M.T.; Yuan, Y.-G. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

History of the Ribosome and the Origin of Translation
History of the ribosome and the origin of translation Anton S. Petrova,1, Burak Gulena, Ashlyn M. Norrisa, Nicholas A. Kovacsa, Chad R. Berniera, Kathryn A. Laniera, George E. Foxb, Stephen C. Harveyc, Roger M. Wartellc, Nicholas V. Huda, and Loren Dean Williamsa,1 aSchool of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, GA 30332; bDepartment of Biology and Biochemistry, University of Houston, Houston, TX, 77204; and cSchool of Biology, Georgia Institute of Technology, Atlanta, GA 30332 Edited by David M. Hillis, The University of Texas at Austin, Austin, TX, and approved November 6, 2015 (received for review May 18, 2015) We present a molecular-level model for the origin and evolution of building up of the functional centers, proceeds to the establishment the translation system, using a 3D comparative method. In this model, of the common core, and continues to the development of large the ribosome evolved by accretion, recursively adding expansion metazoan rRNAs. segments, iteratively growing, subsuming, and freezing the rRNA. Incremental evolution of function is mapped out by stepwise Functions of expansion segments in the ancestral ribosome are accretion of rRNA. In the extant ribosome, specific segments of assigned by correspondence with their functions in the extant rRNA perform specific functions including peptidyl transfer, ribosome. The model explains the evolution of the large ribosomal subunit association, decoding, and energy-driven translocation subunit, the small ribosomal subunit, tRNA, and mRNA. Prokaryotic (11). The model assumes that the correlations of rRNA segments ribosomes evolved in six phases, sequentially acquiring capabilities with their functions have been reasonably maintained over the for RNA folding, catalysis, subunit association, correlated evolution, broad course of ribosomal evolution. -

Structures and Stabilization of Kinetoplastid-Specific Split Rrnas Revealed by Comparing Leishmanial and Human Ribosomes
ARTICLE Received 7 Jun 2016 | Accepted 13 Sep 2016 | Published 18 Oct 2016 DOI: 10.1038/ncomms13223 OPEN Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes Xing Zhang1,2,*, Mason Lai3,*, Winston Chang3, Iris Yu3, Ke Ding3, Jan Mrazek4,HweeL.Ng4, Otto O. Yang2,4, Dmitri A. Maslov5 & Z. Hong Zhou2,3 The recent success in ribosome structure determination by cryoEM has opened the door to defining structural differences between ribosomes of pathogenic organisms and humans and to understand ribosome-targeting antibiotics. Here, by direct electron-counting cryoEM, we have determined the structures of the Leishmania donovani and human ribosomes at 2.9 Å and 3.6 Å, respectively. Our structure of the leishmanial ribosome elucidates the organization of the six fragments of its large subunit rRNA (as opposed to a single 28S rRNA in most eukaryotes, including humans) and reveals atomic details of a unique 20 amino acid extension of the uL13 protein that pins down the ends of three of the rRNA fragments. The structure also fashions many large rRNA expansion segments. Direct comparison of our human and leishmanial ribosome structures at the decoding A-site sheds light on how the bacterial ribosome-targeting drug paromomycin selectively inhibits the eukaryotic L. donovani, but not human, ribosome. 1 Center of Cryo Electron Microscopy, Zhejiang University School of Medicine, Hangzhou, Zhejiang 310058, China. 2 California NanoSystems Institute, University of California, Los Angeles, California 90095, USA. 3 Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, California 90095, USA. 4 Division of Infectious Diseases, Department of Medicine, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, California 90095, USA. -
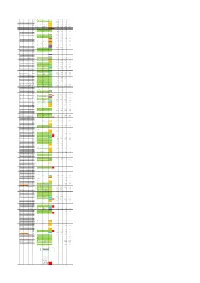
End Strand of Gene Gene Name Gene Function Starnd of Trascript
Genomic TSS strand Gene starnd of Transcipt relatively to TSS Start End Gene name coordinates relatively TSS type Control_fwd TEX_fwd Control_rev TEX_rev of gene function trascript gene group of TSS to ATG ribosomal 93 854 + rps12 + 1 protein S12 1 1 14.0638317 20.7539794 0 0 93 324 exon 1 + 111 rps12 intron 1 1 13.90756687 18.07149224 0.781323982 0.423550599 829 854 exon 2 + 496 rps12 exon2 2 24.22104343 15.24782157 0 0 30S 904 1371 + gene rps7 ribosomal + 2 protein S7 1303 -209 rps7 inter (ndhB) 2 20.47068832 9.035746118 0.625059185 0.847101199 NADH dehydrogen 1512 3535 + gene ndhB (ndh2) + ase subunit 3 2 1696 ndhB exon 1-inter 2 3.594090315 2.964854195 0.468794389 0.282367066 + 2209 ndhB exon 1-inter 2 46.09811492 4.09432246 0.468794389 0.423550599 1512 2237 exon 1 + 2756 ndhB exon 2 -inter 2 43.28534858 4.800240125 0.312529593 0.282367066 2756 3535 exon 2 + 3090 ndhB exon 2 -inter 2 17.50165719 15.95373924 0.312529593 0 + 3192 ndhB exon 2 -inter 2 140.6383167 117.6058831 2.812766334 1.694202397 - 3462 ndhB exon 2 -inter 3 1.406383167 1.129468265 1.406383167 3.67077186 4 3633 3712 + tRNA tRNA-Leu-CAA + 3610 -23 tRNA-Leu 1 77.19480938 84.85130339 0.625059185 0 + 3633 1 tRNA-Leu 1 359.5652963 649.0207016 0.781323982 0 photosyste 3954 4058 + gene psbM m II protein + 5 M 3775 -179 psbM 2 20.47068832 12.00060031 0 0.141183533 + 3954 psbM-0 2 69.22530477 28.37789015 0.156264796 0 hypothetical 4182 5073 + gene ycf66 6 protein 4182 4287 exon 1 4772 5073 exon 2 7 5202 5113 - gene ycf (ORF29) - 5299 orf29 inter 1 0 0 3.125295926 3.67077186 -

HUMAN RIBOSOME BIOGENESIS and the REGULATION of the TUMOUR SUPPRESSOR P53
HUMAN RIBOSOME BIOGENESIS AND THE REGULATION OF THE TUMOUR SUPPRESSOR p53 Andria Pelava Submitted for Doctor of Philosophy Final submission: December 2016 Institute of Cell and Molecular Biosciences Faculty of Medical Sciences Newcastle University ii Abstract Ribosome production is an energetically expensive and, therefore, highly regulated process. Defects in ribosome biogenesis lead to genetic diseases called Ribosomopathies, such as Dyskeratosis Congenita (DC), and mutations in ribosomal proteins and ribosome biogenesis factors are linked to multiple types of cancer. During ribosome biogenesis, the ribosomal RNAs (rRNAs) are processed and modified, and defects in ribosome biogenesis lead to the activation of p53. This project aimed to investigate the functions of Dyskerin, mutated in X-linked DC, in human ribosome biogenesis and p53 regulation, and to explore the link between ribosome production and p53 homeostasis. Dyskerin is an rRNA pseudouridine synthase and required for telomere maintenance. There is some debate as to whether DC is the result of telomere maintenance or ribosome biogenesis defects. It is shown here that human Dyskerin is required for the production of both LSU and SSU, and knockdown of Dyskerin leads to p53 activation via inhibition of MDM2 via the 5S RNP, an LSU assembly intermediate which accumulates after ribosome biogenesis defects. My data indicate that p53 activation, due to defects in ribosome biogenesis, may contribute to the clinical symptoms seen in patients suffering with DC. In addition, it is shown that defects in early or late stages of SSU or LSU biogenesis, result in activation of p53 via the 5S RNP-MDM2 pathway, and that p53 is induced in less than 12 hours after ribosome biogenesis defects. -

Fanconi Anemia Protein FANCI Functions in Ribosome Biogenesis
Fanconi anemia protein FANCI functions in ribosome biogenesis Samuel B. Sondallea, Simonne Longerichb,1, Lisa M. Ogawab, Patrick Sungb,c, and Susan J. Basergaa,b,c,2 aDepartment of Genetics, Yale School of Medicine, New Haven, CT 06520; bDepartment of Molecular Biophysics and Biochemistry, Yale School of Medicine, New Haven, CT 06520; and cDepartment of Therapeutic Radiology, Yale School of Medicine, New Haven, CT 06520 Edited by Joseph G. Gall, Carnegie Institution of Washington, Baltimore, MD, and approved December 28, 2018 (received for review July 8, 2018) Fanconi anemia (FA) is a disease of DNA repair characterized by bone responsible for the transcription of pre-ribosomal RNA (pre- marrow failure and a reduced ability to remove DNA interstrand rRNA) (33). The clear phenotypic overlap between FA and cross-links. Here, we provide evidence that the FA protein FANCI also bone marrow failure ribosomopathies and the connection between functions in ribosome biogenesis, the process of making ribosomes BRCA1 (FANCS) and RNAPI suggest a possible connection be- that initiates in the nucleolus. We show that FANCI localizes to the tween FA and defects in ribosome biogenesis. nucleolus and is functionally and physically tied to the transcription The process of making ribosomes in eukaryotes, termed ri- of pre-ribosomal RNA (pre-rRNA) and to large ribosomal subunit bosome biogenesis, initiates in the large nonmembrane-bound (LSU) pre-rRNA processing independent of FANCD2. While FANCI is nuclear organelle, the nucleolus (NO) (34, 35). In the NO, tan- known to be monoubiquitinated when activated for DNA repair, we dem repeats of ribosomal DNA (rDNA) (36) are transcribed by find that it is predominantly in the deubiquitinated state in the RNAPI, which generates a long pre-rRNA (37). -

Transcriptomic Regulation of Alternative Phenotypic Trajectories in Embryos of the Annual Killifish Austrofundulus Limnaeus
Portland State University PDXScholar Dissertations and Theses Dissertations and Theses Fall 11-30-2017 Transcriptomic Regulation of Alternative Phenotypic Trajectories in Embryos of the Annual Killifish Austrofundulus limnaeus Amie L. Romney Portland State University Follow this and additional works at: https://pdxscholar.library.pdx.edu/open_access_etds Part of the Biology Commons, and the Genetics and Genomics Commons Let us know how access to this document benefits ou.y Recommended Citation Romney, Amie L., "Transcriptomic Regulation of Alternative Phenotypic Trajectories in Embryos of the Annual Killifish Austrofundulus limnaeus" (2017). Dissertations and Theses. Paper 4033. https://doi.org/10.15760/etd.5917 This Dissertation is brought to you for free and open access. It has been accepted for inclusion in Dissertations and Theses by an authorized administrator of PDXScholar. Please contact us if we can make this document more accessible: [email protected]. Transcriptomic Regulation of Alternative Phenotypic Trajectories in embryos of the Annual Killifish Austrofundulus limnaeus by Amie Lynn Thomas Romney A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biology Dissertation Committee Jason Podrabsky, Chair Suzanne Estes Bradley Buckley Todd Rosenstiel Dirk Iwata-Reuyl Portland State University 2017 © 2017 Amie Lynn Thomas Romney ABSTRACT The Annual Killifish, Austrofundulus limnaeus, survives the seasonal drying of their pond habitat in the form of embryos entering diapause midway through development. The diapause trajectory is one of two developmental phenotypes. Alternatively, individuals can “escape” entry into diapause and develop continuously until hatching. The alternative phenotypes of A. limnaeus are a form of developmental plasticity that provides this species with a physiological adaption for surviving stressful environments. -

Ribosomal RNA
Ribosomal RNA Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins.[1] Ribosomal RNA Three-dimensional views of the ribosome, showing rRNA in dark blue (small subunit) is the predominant form of RNA found in most cells; it makes and dark red (large subunit). Lighter colors up about 80% of cellular RNA despite never being translated represent ribosomal proteins. into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass. Contents Structure Assembly Function Subunits and associated ribosomal RNA In prokaryotes In eukaryotes Biosynthesis In eukaryotes Eukaryotic regulation In prokaryotes Prokaryotic regulation Degradation In eukaryotes In prokaryotes Sequence conservation and stability Significance Human genes See also References External links Structure Although the primary structure of rRNA sequences can vary across organisms, base-pairing within these sequences commonly forms stem-loop configurations. The length and position of these rRNA stem-loops allow them to create three-dimensional rRNA structures that are similar across species.[2] Because of these configurations, rRNA can form tight and specific interactions with ribosomal proteins to form ribosomal subunits. These ribosomal proteins contain basic residues (as opposed to acidic residues) and aromatic residues (i.e. -

Supersized Ribosomal RNA Expansion Segments in Asgard Archaea 2 Authors: Petar I
bioRxiv preprint doi: https://doi.org/10.1101/2019.12.25.888164; this version posted June 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Supersized ribosomal RNA expansion segments in Asgard archaea 2 Authors: Petar I. Penev1,2, Sara Fakhretaha-Aval1,3, Vaishnavi J. Patel4, Jamie J. Cannone4, 3 Robin R. Gutell4, Anton S. Petrov1,3*, Loren Dean Williams1,2,3*, Jennifer B. Glass1,2,5* 4 Affiliations: 1NASA Center for the Origin of Life, Georgia Institute of Technology, Atlanta, GA 30332-0400, USA 2School of Biological Sciences, Georgia Institute of Technology, North Avenue, Atlanta, GA 30332, USA 3School of Chemistry and Biochemistry, Georgia Institute of Technology, 901 Atlantic Dr, Atlanta, GA 30332, USA 5 4Department of Integrative Biology, The University of Texas at Austin, 2415 Speedway #C0930, 6 Austin, TX 78712, USA 7 5School of Earth and Atmospheric Sciences, Georgia Institute of Technology, 311 Ferst Dr, 8 Atlanta, GA 30332, USA 9 *Correspondence to: [email protected]; [email protected]; 10 [email protected] 11 1 bioRxiv preprint doi: https://doi.org/10.1101/2019.12.25.888164; this version posted June 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license.