Unraveling the Impact of Bioinformatics and Omics in Agriculture
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Uila Supported Apps
Uila Supported Applications and Protocols updated Oct 2020 Application/Protocol Name Full Description 01net.com 01net website, a French high-tech news site. 050 plus is a Japanese embedded smartphone application dedicated to 050 plus audio-conferencing. 0zz0.com 0zz0 is an online solution to store, send and share files 10050.net China Railcom group web portal. This protocol plug-in classifies the http traffic to the host 10086.cn. It also 10086.cn classifies the ssl traffic to the Common Name 10086.cn. 104.com Web site dedicated to job research. 1111.com.tw Website dedicated to job research in Taiwan. 114la.com Chinese web portal operated by YLMF Computer Technology Co. Chinese cloud storing system of the 115 website. It is operated by YLMF 115.com Computer Technology Co. 118114.cn Chinese booking and reservation portal. 11st.co.kr Korean shopping website 11st. It is operated by SK Planet Co. 1337x.org Bittorrent tracker search engine 139mail 139mail is a chinese webmail powered by China Mobile. 15min.lt Lithuanian news portal Chinese web portal 163. It is operated by NetEase, a company which 163.com pioneered the development of Internet in China. 17173.com Website distributing Chinese games. 17u.com Chinese online travel booking website. 20 minutes is a free, daily newspaper available in France, Spain and 20minutes Switzerland. This plugin classifies websites. 24h.com.vn Vietnamese news portal 24ora.com Aruban news portal 24sata.hr Croatian news portal 24SevenOffice 24SevenOffice is a web-based Enterprise resource planning (ERP) systems. 24ur.com Slovenian news portal 2ch.net Japanese adult videos web site 2Shared 2shared is an online space for sharing and storage. -

Workbench 16 Pgs.PGS
Workbench January 2008 Issue 246 HappyHappy NewNew YearYear AMIGANSAMIGANS 2008 January 2008 Workbench 1 Editorial Happy New Year Folks! Welcome to the first PDF issue of Workbench for 2008. Editor I hope you’ve all had a great Christmas and survived the heat and assorted Barry Woodfield Phone: 9917 2967 weird weather we’ve been having. Mobile : 0448 915 283 I see that YAM is still going strong, having just released Ver. 2.5. Well [email protected] ibutions done, Team. We have a short article on the 25th Anniversary of the C=64 on Contributions can be soft copy (on floppy½ disk) or page four which may prove interesting to hard copy. It will be returned some of you and a few bits of assorted if requested and accompanied with a self- Amiga news on page ten. addressed envelope. Enjoy! The editor of the Amiga Users Group Inc. newsletter Until next month. Ciao for now, Workbench retains the right to edit contributions for Barry R. Woodfield. clarity and length. Send contributions to: Amiga Users Group P.O. Box 2097 Seaford Victoria 3198 OR [email protected] Advertising Advertising space is free for members to sell private items or services. For information on commercial rates, contact: Tony Mulvihill 0415 161 2721 [email protected] Deadlines Last Months Meeting Workbench is published each month. The deadline for each December 9th 2007 issue is the 1st Tuesday of A very good pre-Christmas Gather to the month of publication. Reprints round off the year. All articles in Workbench are Copyright 2007 the Amiga Users Group Inc. -
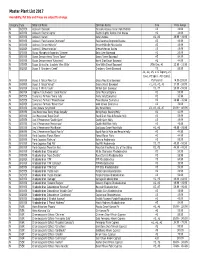
Master Plant List 2017.Xlsx
Master Plant List 2017 Availability, Pot Size and Prices are subject to change. Category Type Botanical Name Common Name Size Price Range N BREVER Azalea X 'Cascade' Cascade Azalea (Glenn Dale Hybrid) #3 49.99 N BREVER Azalea X 'Electric Lights' Electric Lights Double Pink Azalea #2 44.99 N BREVER Azalea X 'Karen' Karen Azalea #2, #3 39.99 - 49.99 N BREVER Azalea X 'Poukhanense Improved' Poukhanense Improved Azalea #3 49.99 N BREVER Azalea X 'Renee Michelle' Renee Michelle Pink Azalea #3 49.99 N BREVER Azalea X 'Stewartstonian' Stewartstonian Azalea #3 49.99 N BREVER Buxus Microphylla Japonica "Gregem' Baby Gem Boxwood #2 29.99 N BREVER Buxus Sempervirens 'Green Tower' Green Tower Boxwood #5 64.99 N BREVER Buxus Sempervirens 'Katerberg' North Star Dwarf Boxwood #2 44.99 N BREVER Buxus Sinica Var. Insularis 'Wee Willie' Wee Willie Dwarf Boxwood Little One, #1 13.99 - 21.99 N BREVER Buxus X 'Cranberry Creek' Cranberry Creek Boxwood #3 89.99 #1, #2, #5, #15 Topiary, #5 Cone, #5 Spiral, #10 Spiral, N BREVER Buxus X 'Green Mountain' Green Mountain Boxwood #5 Pyramid 14.99-299.99 N BREVER Buxus X 'Green Velvet' Green Velvet Boxwood #1, #2, #3, #5 17.99 - 59.99 N BREVER Buxus X 'Winter Gem' Winter Gem Boxwood #5, #7 59.99 - 99.99 N BREVER Daphne X Burkwoodii 'Carol Mackie' Carol Mackie Daphne #2 59.99 N BREVER Euonymus Fortunei 'Ivory Jade' Ivory Jade Euonymus #2 35.99 N BREVER Euonymus Fortunei 'Moonshadow' Moonshadow Euonymus #2 29.99 - 35.99 N BREVER Euonymus Fortunei 'Rosemrtwo' Gold Splash Euonymus #2 39.99 N BREVER Ilex Crenata 'Sky Pencil' -

2021 AAPT Virtual Winter Meeting
2021 AAPT Virtual Winter Meeting VIRTUAL WINTER MEETING 2021 January 9 -12 ® Meet Graphical Analysis Pro We reimagined our award‑winning Vernier Graphical Analysis™ app to help you energize your virtual teaching with real, hands‑on physics. Perfect for Remote Learning • Perform live physics experiments using Vernier sensors and share the data with students in real time. • Create your own videos—synced with actual data—and distribute to students easily. • Explore sample experiments with data that cover important physics topics. Sign up for a free 30-day trial vernier.com/ga-pro-tpt Now offering free webinars & whitepapers from industry leaders Stay connected with the leader in physics news Sign Up to be alerted when new resources become available at physicstoday.org/wwsignup Achieve More in Physics with Macmillan Learning NEW FROM PRINCETON From Nobel Prize–winning Quantum physicist, New York The essential primer for A pithy yet deep introduction physicist P. J. E. Peebles, the Times bestselling author, and physics students who want to to Einstein’s general theory of story of cosmology from BBC host Jim Al-Khalili build their physical intuition relativity Einstein to today offers an illuminating look at Hardcover $35.00 what physics reveals about Hardcover $45.00 Paperback $14.95 the world Hardcover $16.95 Visit our virtual booth SAVE 30% with coupon code APT21 at press.princeton.edu JANUARY 9, 2021 | 12:00 PM - 1:15 PM A1.01 | 21st Century Physics in the Physics Classroom Page 1 A1.02 | Effective Practices in Educational Technology Page -

Amigaos4 Download
Amigaos4 download click here to download Read more, Desktop Publishing with PageStream. PageStream is a creative and feature-rich desktop publishing/page layout program available for AmigaOS. Read more, AmigaOS Application Development. Download the Software Development Kit now and start developing native applications for AmigaOS. Read more.Where to buy · Supported hardware · Features · SDK. Simple DirectMedia Layer port for AmigaOS 4. This is a port of SDL for AmigaOS 4. Some parts were recycled from older SDL port for AmigaOS 4, such as audio and joystick code. Download it here: www.doorway.ru Thank you James! 19 May , In case you haven't noticed yet. It's possible to upload files to OS4Depot using anonymous FTP. You can read up on how to upload and create the required readme file on this page. 02 Apr , To everyone downloading the Diablo 3 archive, April Fools on. File download command line utility: http, https and ftp. Arguments: URL/A,DEST=DESTINATION=TARGET/K,PORT/N,QUIET/S,USER/K,PASSWORD/K,LIST/S,NOSIZE/S,OVERWRITE/S. URL = Download address DEST = File name / Destination directory PORT = Internet port number QUIET = Do not display progress bar. AmigaOS 4 is a line of Amiga operating systems which runs on PowerPC microprocessors. It is mainly based on AmigaOS source code developed by Commodore, and partially on version developed by Haage & Partner. "The Final Update" (for OS version ) was released on 24 December (originally released Latest release: Final Edition Update 1 / De. Purchasers get a serial number inside their box or by email to register their purchase at our website in order to get access to our restricted download area for the game archive, the The game was originally released in for AmigaOS 68k/WarpOS and in December for AmigaOS 4 by Hyperion Entertainment CVBA. -
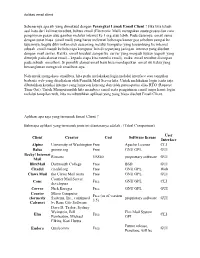
Sebenarnya Apa Sih Yang Dimaksud Dengan Perangkat Lunak Email Client
Aplikasi email client Sebenarnya apa sih yang dimaksud dengan Perangkat Lunak Email Client ? Jika kita telaah asal kata dari kalimat tersebut, bahwa email (Electronic Mail) merupakan suatu proses dan cara pengiriman pesan atau gambar melalui internet ke 1 org atau lebih. Pada dasarnya email sama dengan surat biasa (snail mail) yang harus melewati beberapa kantor pos sebelum sampai ke tujuannya, begitu dikirimkan oleh seseorang melalui komputer yang tersambung ke internet sebuah email masuk ke beberapa komputer lain di sepanjang jaringan internet yang disebut dengan mail server. Ketika email tersebut sampai ke server yang menjadi tujuan (seperti yang ditunjuk pada alamat email – kepada siapa kita menulis email), maka email tersebut disimpan pada sebuah emailbox. Si pemilik alamat email baru bisa mendapatkan email itu kalau yang bersangkutan mengecek emailbox-nya. Nah untuk mengakses emailbox, kita perlu melakukan login melalui interface atau tampilan berbasis web yang disediakan oleh Pemilik Mail Server kita. Untuk melakukan login tentu saja dibutuhkan koneksi internet yang lumayan kencang dan tidak putus-putus alias RTO (Request Time Out). Untuk Mempermudah kita membaca email serta pengiriman email tanpa harus login melalui tampilan web, kita membutuhkan aplikasi yang yang biasa disebut Email Client. Aplikasi apa saja yang termasuk Email Client ? Beberapa aplikasi yang termasuk jenis ini diantaranya adalah : (Tabel Comparison) User Client Creator Cost Software license Interface Alpine University of Washington Free Apache License CLI Balsa gnome.org Free GNU GPL GUI Becky! Internet Rimarts US$40 proprietary software GUI Mail BlitzMail Dartmouth College Free BSD GUI Citadel citadel.org Free GNU GPL Web Claws Mail the Claws Mail team Free GNU GPL GUI Courier Mail Server Cone Free GNU GPL CLI developers Correo Nick Kreeger Free GNU GPL GUI Courier Micro Computer Free (as of version (formerly Systems, Inc., continued proprietary software GUI 3.5) Calypso) by Rose City Software Dave D. -

BALOO's BUGLE Volume 19, Number 8 “Make No Little Plans; They Have No Magic to Stir Men's Blood and Probably Themselves Will Not Be Realized
BALOO'S BUGLE Volume 19, Number 8 “Make no little plans; they have no magic to stir men's blood and probably themselves will not be realized. Make big plans; aim high in hope and work." — Daniel Hudson Burnham (1846-1912) ---------------------------------------------------------------------------------------------------------------- April 2013 Cub Scout Roundtable May 2013Core Value & Pack Meeting Ideas HEALTH & FITNESS / CUB CAFE Tiger Cub, Wolf, Bear, Webelos, & Arrow of Light Activities So, if you are a RT Commissioner, an Asst RT CORE VALUES Commissioner (formerly called Staff) or just The core value highlighted this month is: interested in RTs - for a great experience, go to Health and Fitness: Being personally committed to http://philmontscoutranch.org learn about the keeping our minds and bodies clean and fit. By participating in the Cub Scout Academics and Sports Philmont Training Center and sign up for Effective program, Cub Scouts and their families develop an Roundtables and then come on out and meet understanding of the benefits of being fit and healthy. George and I and learn a lot about Roundtables. We will have the 2013-2014 CS and BS RT Planning Guides for you!! Write PTC (or me) if you have questions on the National Training Center - This COMMISSIONER’S CORNER will be my 13th trip there - the first on "Phil Phaculty!!" June 23-29 is Commissioner's Week at PTC Commissioner Conferences being offered include The Council Commissioner Table of Contents District Commissioner and Asst Dist Commissioner CORE VALUES ..................................................... 1 Training COMMISSIONER’S CORNER ............................. 1 District Committee Table of Contents .................................................... 1 Effective Roundtables THOUGHTFUL ITEMS FOR SCOUTERS .......... 2 How to Conduct a Commissioner College Prayer ................................................................. -

Dental Exhibition and Congress 12-13-14 October 2017 Budapest – Hungary
www.azadmed.com l&Hazaadtejaratpars C)@azadmed2 \. 88 98 80 63 - 6 All the advertisements you see are companies looking for distributors, contact them now to enlarge your business. Edited by ImplantBook More than 200 products 2-2017 of 153 international app May /July companies. free & easy to use! implant-book.com www.infodent.com INFODENT 2/2017- INFODENT Srl - Str. Cassia Nord Km 86,300 01100 Viterbo - Italy • Help others Visit us at Sino-Dental 2017 Beijing to find you • Get what you are looking for • Engage more potential partners infodent.com 97,54 % FOLLOW-UP from 2010 to 2016 Data collection published during 5th Oralplant Conference - Venice 2016 Short L 4.5 Extra-Short L 2.5 Stay up to date on new implantology solutions. Would you like to be our Distributor? Oralplant Suisse SA The quality system of UNI EN ISO 9001 Oralplant S.r.l. Oralplant S.r.l. EN ISO 13485 [email protected] has been found to comply EC - CERTIFICATE MDD [email protected] www.oralplant.ch with the requirements of Medical Device Directive www.oralplant.it Sanctuary™ Latex Black Dental Dam HOTOG Color Contrast :-=.::::.- --- -:.=....-- -- - -· :::::=.,.;::- -- ,.,. __ ·--- """•,...., ...... ,_..,. TIW.DE AiSVISOR ++++½ i nfo@san ctua ryhea Ith .com. my sanctuary dental . sanctuary dental cJ_ www .sanctuary-dental.com EDITORIAL Digital, Smart, Patient-Oriented For Dealership Opportunity: • [email protected] DIGITAL, SMART, PATIENT-ORIENTED • www.beyes.ca The leading global trade fair of the dental What is striking, for instance, is that advanced economies enjoy The fastest growing Canadian dental company industry, the IDS 2017, closed its doors last large economic benefits from digitization, such as growth and pro- March with the focus laying on digital pro- ductivity, but gain less in terms of jobs. -

What,To Do Regarding Eapnomics and Managing Ohio State Dept
DOCUMENT RJSUME ED 259 182 CE 041 904 TITLE ,What,to Do Regarding Eapnomics and Managing Resources. INSTITUTION Ohio State Dept. of Education, Columbus. Div. of Vocational Education.; Ohio State Univ., Columbus. Instructional Materials Lab. PUB DATE Aug 83 t) NOTE 350p.; For related documents, see CE 041 900-906. PUB TYPE* Guides Classroom Use Guides (For Teachers) (052) A EDRS PRIC MF01/PC14 Plus Postage. DESCRIPT S Behavioral Objectives; Citizen Participation; *Consumer Economics; *Consumer Education; *Consumer Protection; Curriculum Guides; Decision Making; Family Life Education; *Home Econqmics; *Homemaking Skills; Home Management; Learning Activities; Learning Modules; *Money Management; Secondary Education ABSTRACT These materials for the curriculum area of economics and managing resources comprise one of six such'packages that are part of the Ohio Vocational.Consumer/Homemaking Curriculum Guide. The curriculum area or perennial problem taken up in this document is ._divided into three practical problems about what to do regarding: (1) decision making; (2) resource/product management; and (3) citizen participation. These are further categorized intro seven concerns: external and personal factors affecting consumer decisions, financia1. planning, purchasing, conserving, consumer protection, and consumer responsibilities. Each concern is divided into a number of concepts * or modules. This package consists of 25 modules. Theformat for each module is as follows: code, perennial problem, practical problem, concern/concept, homemaking skills (listing of various skills needed by the homemaker as related to the ideveloped.concepts), and a chart relating process skills (steps of practical reasoning), concepts (further breakdown of the topic), And strategies (information and activities that facilitate the teaching/learning of the concepts). -
Download Issue 3
CONTENTS CONTENTS Contents The Chairman News Speaks Editorial UPDATE SEAL Update...................... 3 irstly I have an apology to make, id you know that brainwashing is News Items......................... 4 as Im sure youve noticed this totally legal?... Let me tell you a Members Amiga Update..................... 7 Fissue is very late, due to several Dstory. Since the last magazine we’ve helped other commitments (SEAL, WoA and several more SEAL members get on the work related) Ive been unable to find the I have been into photography for some it out yourself. I haven’t got a computer Internet and we hope this issue will en- time I usually do to work on the maga- time now, and had decided it was time to myself, so I don’t know which ones it will courage even more people to get con- Features zine. However now its here I hope youll enter into the world of digital photog- work on (Dixons well-trained staff, know- nected. Members will find the latest find this is our best issue yet. We have raphy, as the cameras are now more ledgeable and as sharp as ever). members list complete with EMail ad- Interview with Petro ............ 8 more pages (up from 36 to 40) and affordable and have better specifications dresses printed on the back of the Get Netted! ......................... 10 much better quality printing thanks to than earlier models. Once I had So I bought the camera and was chuffed covering letter accompanying their mag- Choosing An ISP ................ 14 our new printers who print from researched into which drivers were with myself (I’ve been after one for a azine. -

Case Studies from Developing Countries in Crops, Livestock and Fish Occasional Papers on Innovation in Family Farming
OCCASIONAL PAPERS ON INNOVATION IN FAMILY FARMING BIOTECHNOLOGIES AT WORK FOR SMALLHOLDERS: CASE STUDIES FROM DEVELOPING COUNTRIES IN CROPS, LIVESTOCK AND FISH OCCASIONAL PAPERS ON INNOVATION IN FAMILY FARMING BIOTECHNOLOGIES AT WORK FOR SMALLHOLDERS: CASE STUDIES FROM DEVELOPING COUNTRIES IN CROPS, LIVESTOCK AND FISH Edited by J. Ruane, J.D. Dargie, C. Mba, P. Boettcher, H.P.S. Makkar, D.M. Bartley and A. Sonnino Food and Agriculture Organization of the United Nations 2013 The designations employed and the presentation of material in this information product do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations (FAO) concerning the legal or development status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. The mention of specific companies or products of manufacturers, whether or not these have been patented, does not imply that these have been endorsed or recommended by FAO in preference to others of a similar nature that are not mentioned. The views expressed in this information product are those of the author(s) and do not necessarily reflect the views or policies of FAO. ISBN 978-92-5-107877-8 (print) E-ISBN 978-92-5-107878-5 (PDF) © FAO, 2013 FAO encourages the use, reproduction and dissemination of material in this information product. Except where otherwise indicated, material may be copied, downloaded and printed for private study, research and teaching purposes, or for use in non-commercial products or services, provided that appropriate acknowledgement of FAO as the source and copyright holder is given and that FAO’s endorsement of users’ views, products or services is not implied in any way. -

Caesarstone Sdot-Yam Ltd
CAESARSTONE SDOT-YAM LTD. FORM 20-F (Annual and Transition Report (foreign private issuer)) Filed 05/13/14 for the Period Ending 12/31/13 Telephone 972 4 636 4555 CIK 0001504379 Symbol CSTE SIC Code 3281 - Cut Stone and Stone Products Industry Constr. - Supplies & Fixtures Sector Capital Goods http://www.edgar-online.com © Copyright 2014, EDGAR Online, Inc. All Rights Reserved. Distribution and use of this document restricted under EDGAR Online, Inc. Terms of Use. UNITED STATES SECURITIES AND EXCHANGE COMMISSION Washington, D.C. 20549 Form 20-F (Mark One) REGISTRATION STATEMENT PURSUANT TO SECTION 12(b) OR (g) OF THE SECURITIES EXCHANGE ACT OF 1934 OR ANNUAL REPORT PURSUANT TO SECTION 13 OR 15(d) OF THE SECURITIES EXCHANGE ACT OF 1934 For the fiscal year ended December 31, 2013 OR TRANSITION REPORT PURSUANT TO SECTION 13 OR 15(d) OF THE SECURITIES EXCHANGE ACT OF 1934 For the transition period from ______ to ______ OR SHELL COMPANY REPORT PURSUANT TO SECTION 13 or 15(d) OF THE SECURITIES EXCHANGE ACT OF 1934 Date of event requiring this shell company report…………………………………. Commission File Number 001-35464 CAESARSTONE SDOT-YAM LTD. (Exact Name of Registrant as specified in its charter) ISRAEL (Jurisdiction of incorporation or organization) Kibbutz Sdot-Yam MP Menashe, 3780400 Israel (Address of principal executive offices) Yosef Shiran Chief Executive Officer Caesarstone Sdot-Yam Ltd. MP Menashe, 3780400 Israel Telephone: +972 (4) 636-4555 Fascimile: +972 (4) 636-4400 (Name, telephone, email and/or facsimile number and address of company