Association of the Serotonin Receptor 3E Gene As a Functional Variant In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Research Article Microarray-Based Comparisons of Ion Channel Expression Patterns: Human Keratinocytes to Reprogrammed Hipscs To
Hindawi Publishing Corporation Stem Cells International Volume 2013, Article ID 784629, 25 pages http://dx.doi.org/10.1155/2013/784629 Research Article Microarray-Based Comparisons of Ion Channel Expression Patterns: Human Keratinocytes to Reprogrammed hiPSCs to Differentiated Neuronal and Cardiac Progeny Leonhard Linta,1 Marianne Stockmann,1 Qiong Lin,2 André Lechel,3 Christian Proepper,1 Tobias M. Boeckers,1 Alexander Kleger,3 and Stefan Liebau1 1 InstituteforAnatomyCellBiology,UlmUniversity,Albert-EinsteinAllee11,89081Ulm,Germany 2 Institute for Biomedical Engineering, Department of Cell Biology, RWTH Aachen, Pauwelstrasse 30, 52074 Aachen, Germany 3 Department of Internal Medicine I, Ulm University, Albert-Einstein Allee 11, 89081 Ulm, Germany Correspondence should be addressed to Alexander Kleger; [email protected] and Stefan Liebau; [email protected] Received 31 January 2013; Accepted 6 March 2013 Academic Editor: Michael Levin Copyright © 2013 Leonhard Linta et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Ion channels are involved in a large variety of cellular processes including stem cell differentiation. Numerous families of ion channels are present in the organism which can be distinguished by means of, for example, ion selectivity, gating mechanism, composition, or cell biological function. To characterize the distinct expression of this group of ion channels we have compared the mRNA expression levels of ion channel genes between human keratinocyte-derived induced pluripotent stem cells (hiPSCs) and their somatic cell source, keratinocytes from plucked human hair. This comparison revealed that 26% of the analyzed probes showed an upregulation of ion channels in hiPSCs while just 6% were downregulated. -

Ion Channels
UC Davis UC Davis Previously Published Works Title THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels. Permalink https://escholarship.org/uc/item/1442g5hg Journal British journal of pharmacology, 176 Suppl 1(S1) ISSN 0007-1188 Authors Alexander, Stephen PH Mathie, Alistair Peters, John A et al. Publication Date 2019-12-01 DOI 10.1111/bph.14749 License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology (2019) 176, S142–S228 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels Stephen PH Alexander1 , Alistair Mathie2 ,JohnAPeters3 , Emma L Veale2 , Jörg Striessnig4 , Eamonn Kelly5, Jane F Armstrong6 , Elena Faccenda6 ,SimonDHarding6 ,AdamJPawson6 , Joanna L Sharman6 , Christopher Southan6 , Jamie A Davies6 and CGTP Collaborators 1School of Life Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK 2Medway School of Pharmacy, The Universities of Greenwich and Kent at Medway, Anson Building, Central Avenue, Chatham Maritime, Chatham, Kent, ME4 4TB, UK 3Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK 4Pharmacology and Toxicology, Institute of Pharmacy, University of Innsbruck, A-6020 Innsbruck, Austria 5School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, BS8 1TD, UK 6Centre for Discovery Brain Science, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2019/20 is the fourth in this series of biennial publications. The Concise Guide provides concise overviews of the key properties of nearly 1800 human drug targets with an emphasis on selective pharmacology (where available), plus links to the open access knowledgebase source of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. -

Structural Basis for Functional Modulation of Pentameric Ligand-Gated Ion Channels
STRUCTURAL BASIS FOR FUNCTIONAL MODULATION OF PENTAMERIC LIGAND-GATED ION CHANNELS by YVONNE W. GICHERU Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Thesis Advisor: Sudha Chakrapani, Ph.D. Department of Physiology and Biophysics CASE WESTERN RESERVE UNIVERSITY May 2019 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of YVONNE W. GICHERU Candidate for the degree of Physiology and Biophysics* Witold Surewicz (Committee Chair) Matthias Buck Stephen Jones Vera Moiseenkova-Bell Rajesh Ramachandran Sudha Chakrapani March 27, 2019 *We also certify that written approval has been obtained for any proprietary material contained therein. Dedication To my family, friends, mentors, and all who have supported me through this process, thank you. Table of Contents List of Figures .................................................................................................... iv List of Abbreviations .......................................................................................... v Abstract .............................................................................................................. vi Chapter 1 ............................................................................................................. 1 Introduction .................................................................................................... 1 1.1 Pentameric ligand-gated ion channel (pLGIC) superfamily ...................... 2 1.2 pLGIC architecture -

A Bioinformatics Model of Human Diseases on the Basis Of
SUPPLEMENTARY MATERIALS A Bioinformatics Model of Human Diseases on the basis of Differentially Expressed Genes (of Domestic versus Wild Animals) That Are Orthologs of Human Genes Associated with Reproductive-Potential Changes Vasiliev1,2 G, Chadaeva2 I, Rasskazov2 D, Ponomarenko2 P, Sharypova2 E, Drachkova2 I, Bogomolov2 A, Savinkova2 L, Ponomarenko2,* M, Kolchanov2 N, Osadchuk2 A, Oshchepkov2 D, Osadchuk2 L 1 Novosibirsk State University, Novosibirsk 630090, Russia; 2 Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences, Novosibirsk 630090, Russia; * Correspondence: [email protected]. Tel.: +7 (383) 363-4963 ext. 1311 (M.P.) Supplementary data on effects of the human gene underexpression or overexpression under this study on the reproductive potential Table S1. Effects of underexpression or overexpression of the human genes under this study on the reproductive potential according to our estimates [1-5]. ↓ ↑ Human Deficit ( ) Excess ( ) # Gene NSNP Effect on reproductive potential [Reference] ♂♀ NSNP Effect on reproductive potential [Reference] ♂♀ 1 increased risks of preeclampsia as one of the most challenging 1 ACKR1 ← increased risk of atherosclerosis and other coronary artery disease [9] ← [3] problems of modern obstetrics [8] 1 within a model of human diseases using Adcyap1-knockout mice, 3 in a model of human health using transgenic mice overexpressing 2 ADCYAP1 ← → [4] decreased fertility [10] [4] Adcyap1 within only pancreatic β-cells, ameliorated diabetes [11] 2 within a model of human diseases -

The Characterization of Cell Surface Receptor Complexes by Affinity Chromatography, Liquid Chromatography and Tandem Mass Spectrometry
The Characterization of Cell Surface Receptor Complexes by Affinity Chromatography, Liquid Chromatography and Tandem Mass Spectrometry by Jaimie Dufresne BSc, MSc, Ryerson University A dissertation presented to Ryerson University in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Program of Molecular Science Toronto, Ontario, Canada, 2017 © Jaimie Dufresne 2017 AUTHOR'S DECLARATION FOR ELECTRONIC SUBMISSION OF A DISSERTATION I hereby declare that I am the sole author of this dissertation. This is a true copy of the dissertation, including any required final revisions, as accepted by my examiners. I authorize Ryerson University to lend this dissertation to other institutions or individuals for the purpose of scholarly research. I further authorize Ryerson University to reproduce this dissertation by photocopying or by other means, in total or in part, at the request of other institutions or individuals for the purpose of scholarly research. I understand that my dissertation may be made electronically available to the public. ii Abstract Cell surface receptors are of critical importance to the treatment of disease but are difficult to isolate and identify by classical approaches. Here, a robust and general method for capturing a receptor complex from the surface of live cells with ligands presented on nanoscopic beads is demonstrated. Two forms of affinity chromatography: the presentation of a biotinylated ligand to the surface of live cells and recovered by classical affinity chromatography was compared to the presentation of the ligand on the surface of nanoscopic chromatography beads for the isolation of the IgG-FcR complex from the surface of live cells. -

Output Results of CLIME (Clustering by Inferred Models of Evolution)
Output results of CLIME (CLustering by Inferred Models of Evolution) Dataset: Num of genes in input gene set: 9 Total number of genes: 20834 Prediction LLR threshold: 0 The CLIME PDF output two sections: 1) Overview of Evolutionarily Conserved Modules (ECMs) Top panel shows the predefined species tree. Bottom panel shows the partition of input genes into Evolutionary Conserved Modules (ECMs), ordered by ECM strength (shown at right), and separated by horizontal lines. Each row show one gene, where the phylogenetic profile indicates presence (blue) or absence (gray) of homologs in each species (column). Gene symbols are shown at left. Gray color indicates that the gene is a paralog to a higher scoring gene within the same ECM (based on BLASTP E < 1e-3). 2) Details of each ECM and its expansion ECM+ Top panel shows the inferred evolutionary history on the predefined species tree. Branch color shows the gain event (blue) and loss events (red color, with brighter color indicating higher confidence in loss). Branches before the gain or after a loss are shown in gray. Bottom panel shows the input genes that are within the ECM (blue/white rows) as well as all genes in the expanded ECM+ (green/gray rows). The ECM+ includes genes likely to have arisen under the inferred model of evolution relative to a background model, and scored using a log likelihood ratio (LLR). PG indicates "paralog group" and are labeled alphabetically (i.e., A, B). The first gene within each paralog group is shown in black color. All other genes sharing sequence similarity (BLAST E < 1e-3) are assigned to the same PG label and displayed in gray. -

An Enigmatic Case of Cardiac Death in an 18-Years Old Girl
European Review for Medical and Pharmacological Sciences 2021; 25: 4999-5005 An enigmatic case of cardiac death in an 18-years old girl G.A. LANZA1, M. COLL2, S. GRASSI3, V. ARENA4, O. CAMPUZANO5, L. CALÒ6, E. DE RUVO6, R. BRUGADA7, A. OLIVA3 1Department of Cardiovascular Sciences, Fondazione Policlinico A. Gemelli IRCCS, Università Cattolica del Sacro Cuore, Roma, Italy 2Cardiovascular Genetics Centre, University of Girona, Girona, Spain 3Department of Health Surveillance and Bioethics, Fondazione Policlinico A. Gemelli IRCCS, Università Cattolica del Sacro Cuore, Roma, Italy 4Department of Woman and Child Health and Public Health, Fondazione Policlinico A. Gemelli IRCCS, Università Cattolica del Sacro Cuore, Roma, Italy 5Centro Investigación Biomédica en Red Enfermedades Cardiovasculares (CIBERCV), Madrid, Spain 6Division of Cardiology, Policlinico Casilino, ASL Rome B, Roma, Italy 7Department of Medical Science, University of Girona, Girona, Spain Abstract. – We report a case of unusual and of death related to severe abnormalities of the unexplained cardiac death in an 18-years old electrical activation of the heart, eventually re- female patient with congenital neurosensori- sulting in hemodynamic compromise and death. al deafness. The fatal event was characterized by an initial syncopal episode, associated with Clinical Case a wide QRS tachycardia (around 110 bpm) but stable hemodynamic conditions. The patient, An 18-years old girl was referred to the Emer- however, subsequently developed severe hypo- gency Department (ED) of a hospital in Rome, tension and progressive bradyarrhythmias until Italy, after a syncopal episode, occurring while asystole and lack of cardiac response to resus- she was at school. Syncope was preceded by sub- citation maneuvers and ventricular pacing. -

The Hypothalamus As a Hub for SARS-Cov-2 Brain Infection and Pathogenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The hypothalamus as a hub for SARS-CoV-2 brain infection and pathogenesis Sreekala Nampoothiri1,2#, Florent Sauve1,2#, Gaëtan Ternier1,2ƒ, Daniela Fernandois1,2 ƒ, Caio Coelho1,2, Monica ImBernon1,2, Eleonora Deligia1,2, Romain PerBet1, Vincent Florent1,2,3, Marc Baroncini1,2, Florence Pasquier1,4, François Trottein5, Claude-Alain Maurage1,2, Virginie Mattot1,2‡, Paolo GiacoBini1,2‡, S. Rasika1,2‡*, Vincent Prevot1,2‡* 1 Univ. Lille, Inserm, CHU Lille, Lille Neuroscience & Cognition, DistAlz, UMR-S 1172, Lille, France 2 LaBoratorY of Development and PlasticitY of the Neuroendocrine Brain, FHU 1000 daYs for health, EGID, School of Medicine, Lille, France 3 Nutrition, Arras General Hospital, Arras, France 4 Centre mémoire ressources et recherche, CHU Lille, LiCEND, Lille, France 5 Univ. Lille, CNRS, INSERM, CHU Lille, Institut Pasteur de Lille, U1019 - UMR 8204 - CIIL - Center for Infection and ImmunitY of Lille (CIIL), Lille, France. # and ƒ These authors contriButed equallY to this work. ‡ These authors directed this work *Correspondence to: [email protected] and [email protected] Short title: Covid-19: the hypothalamic hypothesis 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

A New Mechanism of Receptor Targeting by Interaction Between Two Classes of Ligand-Gated Ion Channels
1456 • The Journal of Neuroscience, February 3, 2016 • 36(5):1456–1470 Cellular/Molecular A New Mechanism of Receptor Targeting by Interaction between Two Classes of Ligand-Gated Ion Channels Michel Boris Emerit,1,2 Camille Baranowski,1,2 Jorge Diaz,1,2 Audrey Martinez,3,4 Julie Areias,1,2 Jeanine Alterio,1,2 Justine Masson,1,2 XEric Boue´-Grabot,3,4 and XMiche`le Darmon1,2 1Centre de Psychiatrie et Neurosciences, INSERM UMR 894, 75013 Paris, France, 2Universite´ Paris V, 75006 Paris, France, 3Institut des Maladies Neurode´ge´ne´ratives, CNRS UMR 5293, 33076 Bordeaux, France, and 4Universite´ de Bordeaux, 33076 Bordeaux, France The 5-HT3 receptors are serotonin-gated ion channels that physically couple with purinergic P2X2 receptors to trigger a functional cross-inhibition leading to reciprocal channel occlusion. Although this functional receptor–receptor coupling seems to serve a modula- tory role on both channels, this might not be its main physiological purpose. Using primary cultures of rat hippocampal neurons as a quantitative model of polarized targeting, we show here a novel function for this interaction. In this model, 5-HT3A receptors did not exhibit by themselves the capability of distal targeting in dendrites and axons but required the presence of P2X2R for their proper subcellular localization. 5-HT3AR distal targeting occurred with a delayed time course and exhibited a neuron phenotype dependency. In the subpopulation of neurons expressing endogenous P2X2R, 5-HT3AR distal neuritic localization correlated with P2X2R expression and could be selectively inhibited by P2X2R RNA interference. Cotransfection of both receptors revealed a specific colocalization, cotraffick- ing in common surface clusters, and the axonal rerouting of 5-HT3AR. -

HTR3A and HTR3E Gene Polymorphisms and Diarrhea Predominant Irritable Bowel Syndrome Risk: Evidence from a Meta-Analysis
www.impactjournals.com/oncotarget/ Oncotarget, 2017, Vol. 8, (No. 59), pp: 100459-100468 Meta-Analysis HTR3A and HTR3E gene polymorphisms and diarrhea predominant irritable bowel syndrome risk: evidence from a meta-analysis Tangming Guan1,*, Ting Li1,*, Weiming Cai1,2,*, Dong Huang1, Peipei Ouyang3, Yan Wang1, Huayan Chen1, Kefeng Wu4 and Xiaoli Ma1,5 1Department of Pharmacy, Affiliated Hospital of Guangdong Medical University, Zhanjiang, Guangdong Province 524001, China 2Laboratory of Clinical Pharmacy, Guangdong Medical University, Zhanjiang, Guangdong Province 524001, China 3Department of Pharmacology, Guangdong Medical University, Zhanjiang, Guangdong Province 524001, China 4Guangdong Key Laboratory for Research and Development of Natural Drug, Guangdong Medical University, Zhanjiang, Guangdong Province 524001, China 5Department of Clinical Pharmacy, Guangdong Medical University, Zhanjiang, Guangdong Province 524001, China *These authors contributed equally to this work Correspondence to: Kefeng Wu, email: [email protected] Xiaoli Ma, email: [email protected] Keywords: HTR3A, HTR3E, polymorphism, meta-analysis, diarrhea predominant irritable bowel syndrome Received: June 12, 2017 Accepted: July 13, 2017 Published: July 29, 2017 Copyright: Guan et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Several studies have reported an association between serotonin receptor type 3 (5-HT3) subunit genes HTR3A (rs1062613) and HTR3E (rs62625044) and diarrhea predominant irritable bowel syndrome (IBS-D). However, the results remain inconclusive and controversial, particularly for the data derived from different ethnicities and genders. Therefore, we performed a meta-analysis to evaluate this association. -

Ion Channels Expression and Function Are Strongly Modified in Solid Tumors and Vascular Malformations
Biasiotta et al. J Transl Med (2016) 14:285 DOI 10.1186/s12967-016-1038-y Journal of Translational Medicine RESEARCH Open Access Ion channels expression and function are strongly modified in solid tumors and vascular malformations Antonella Biasiotta1†, Daniela D’Arcangelo2†, Francesca Passarelli2, Ezio Maria Nicodemi2* and Antonio Facchiano2* Abstract Background: Several cellular functions relate to ion-channels activity. Physiologically relevant chains of events lead- ing to angiogenesis, cell cycle and different forms of cell death, require transmembrane voltage control. We hypothe- sized that the unordered angiogenesis occurring in solid cancers and vascular malformations might associate, at least in part, to ion-transport alteration. Methods: The expression level of several ion-channels was analyzed in human solid tumor biopsies. Expression of 90 genes coding for ion-channels related proteins was investigated within the Oncomine database, in 25 independ- ent patients-datasets referring to five histologically-different solid tumors (namely, bladder cancer, glioblastoma, melanoma, breast invasive-ductal cancer, lung carcinoma), in a total of 3673 patients (674 control-samples and 2999 cancer-samples). Furthermore, the ion-channel activity was directly assessed by measuring in vivo the electrical sym- pathetic skin responses (SSR) on the skin of 14 patients affected by the flat port-wine stains vascular malformation, i.e., a non-tumor vascular malformation clinical model. Results: Several ion-channels showed significantly increased expression in tumors (p < 0.0005); nine genes (namely, CACNA1D, FXYD3, FXYD5, HTR3A, KCNE3, KCNE4, KCNN4, CLIC1, TRPM3) showed such significant modification in at least half of datasets investigated for each cancer type. Moreover, in vivo analyses in flat port-wine stains patients showed a significantly reduced SSR in the affected skin as compared to the contralateral healthy skin (p < 0.05), in both latency and amplitude measurements. -
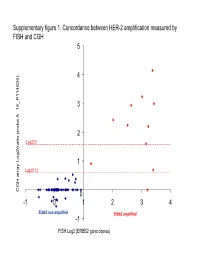
Supplementary Data
Supplementary figure 1. Concordance between HER-2 amplification measured by FISH and CGH 5 4 6) 3 14_P11482 __ 2 Log2(3) atio probe A atio probe rr 1 Log2(1.5) array Log2( CGH 0 -1 0 1 2 3 4 Er bb2 non amplifi e d Erbb2 amplified -1 FISH Log2 (ERBB2 gene copies) Supplementary Figure 2. Gained and lost regions according to triple-negativity Non-triple negative tumors (n=69) Triple negative tumors (n=35) Non-triple negative tumors (n=69) vs triple negative tumors (n=35) Supplementary Figure 3. Gained and lost regions according to chemotherapy response Supplementary Figure 4. Gained and lost regions according to taxanes efficacy Supplementary Table 1. Patient characteristics (n=106). Characteristic N (%) Age (median, range) 51 (28-73) cT cT0/T1 6 (6%) cT2 53 (50%) cT3 18 (17%) cT4 29 (17%) cN cN0 24 (23%) cN1 50 (47%) cN2 23 (22%) cN3 9 (8%) Tumor grade Grade 1 6 (6%) Grade 2 39 (37%) Grade 3 47 (43%) Not assessable 14 (14%) Estrogen receptor expression Negative 45 (42%) Positive 60 (57%) Unknown 1 (1%) Progesteron receptor expression Negative 57 (54%) Positive 47 (44%) Unknown 2 (2%) Her2 status Normal 95 (90%) Overexpression and/or amplification 11 (10%) Preoperative chemotherapy T/FAC* 66 (62%) FAC 38 (36%) Trastuzumab-containing regimen 2 (2%) Pathologic complete remission No 83 (78%) Yes 20 (19%) Not assessable 3 (3%) *: Sequential Paclitaxel / FAC (or FEC) or docetaxel /FEC (n=2) Supplementary Table 2. Minimum Common region (MCR) gained in more than 20% of samples: Band1 Probes Freq Genes AP2M1 CLCN2 DVL3 EIF4G1 EPHB3 POLR2H PSMD2 THPO