MBNL1 and RBFOX2 Cooperate to Establish a Splicing Programme Involved in Pluripotent Stem Cell Differentiation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

SRC Antibody - N-Terminal Region (ARP32476 P050) Data Sheet
SRC antibody - N-terminal region (ARP32476_P050) Data Sheet Product Number ARP32476_P050 Product Name SRC antibody - N-terminal region (ARP32476_P050) Size 50ug Gene Symbol SRC Alias Symbols ASV; SRC1; c-SRC; p60-Src Nucleotide Accession# NM_005417 Protein Size (# AA) 536 amino acids Molecular Weight 60kDa Product Format Lyophilized powder NCBI Gene Id 6714 Host Rabbit Clonality Polyclonal Official Gene Full Name V-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) Gene Family SH2D This is a rabbit polyclonal antibody against SRC. It was validated on Western Blot by Aviva Systems Biology. At Aviva Systems Biology we manufacture rabbit polyclonal antibodies on a large scale (200-1000 Description products/month) of high throughput manner. Our antibodies are peptide based and protein family oriented. We usually provide antibodies covering each member of a whole protein family of your interest. We also use our best efforts to provide you antibodies recognize various epitopes of a target protein. For availability of antibody needed for your experiment, please inquire (). Peptide Sequence Synthetic peptide located within the following region: QTPSKPASADGHRGPSAAFAPAAAEPKLFGGFNSSDTVTSPQRAGPLAGG This gene is highly similar to the v-src gene of Rous sarcoma virus. This proto-oncogene may play a role in the Description of Target regulation of embryonic development and cell growth. SRC protein is a tyrosine-protein kinase whose activity can be inhibited by phosphorylation by c-SRC kinase. Mutations in this gene could be involved in the -

Interplay Between Coding and Exonic Splicing Regulatory Sequences
Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Research Interplay between coding and exonic splicing regulatory sequences Nicolas Fontrodona,1,4 Fabien Aubé,1,4 Jean-Baptiste Claude,1 Hélène Polvèche,1,5 Sébastien Lemaire,1 Léon-Charles Tranchevent,2 Laurent Modolo,3 Franck Mortreux,1 Cyril F. Bourgeois,1 and Didier Auboeuf1 1Université Lyon, ENS de Lyon, Université Claude Bernard, CNRS UMR 5239, INSERM U1210, Laboratory of Biology and Modelling of the Cell, F-69007, Lyon, France; 2Proteome and Genome Research Unit, Department of Oncology, Luxembourg Institute of Health (LIH), L-1445 Strassen, Luxembourg; 3LBMC Biocomputing Center, CNRS UMR 5239, INSERM U1210, F-69007, Lyon, France The inclusion of exons during the splicing process depends on the binding of splicing factors to short low-complexity reg- ulatory sequences. The relationship between exonic splicing regulatory sequences and coding sequences is still poorly un- derstood. We demonstrate that exons that are coregulated by any given splicing factor share a similar nucleotide composition bias and preferentially code for amino acids with similar physicochemical properties because of the nonran- domness of the genetic code. Indeed, amino acids sharing similar physicochemical properties correspond to codons that have the same nucleotide composition bias. In particular, we uncover that the TRA2A and TRA2B splicing factors that bind to adenine-rich motifs promote the inclusion of adenine-rich exons coding preferentially for hydrophilic amino acids that correspond to adenine-rich codons. SRSF2 that binds guanine/cytosine-rich motifs promotes the inclusion of GC-rich exons coding preferentially for small amino acids, whereas SRSF3 that binds cytosine-rich motifs promotes the inclusion of exons coding preferentially for uncharged amino acids, like serine and threonine that can be phosphorylated. -

Splicing Regulatory Factors in Breast Cancer Hallmarks and Disease Progression
www.oncotarget.com Oncotarget, 2019, Vol. 10, (No. 57), pp: 6021-6037 Review Splicing regulatory factors in breast cancer hallmarks and disease progression Esmee Koedoot1, Liesanne Wolters1, Bob van de Water1 and Sylvia E. Le Dévédec1 1Division of Drug Discovery and Safety, LACDR, Leiden University, Leiden, The Netherlands Correspondence to: Sylvia E. Le Dévédec, email: [email protected] Keywords: hallmarks of cancer; breast cancer; alternative splicing; splice factors; RNA sequencing Received: April 23, 2019 Accepted: August 29, 2019 Published: October 15, 2019 Copyright: Koedoot et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT By regulating transcript isoform expression levels, alternative splicing provides an additional layer of protein control. Recent studies show evidence that cancer cells use different splicing events to fulfill their requirements in order to develop, progress and metastasize. However, there has been less attention for the role of the complex catalyzing the complicated multistep splicing reaction: the spliceosome. The spliceosome consists of multiple sub-complexes in total comprising 244 proteins or splice factors and 5 associated RNA molecules. Here we discuss the role of splice factors in the oncogenic processes tumors cells need to fulfill their oncogenic properties (the so-called the hallmarks of cancer). Despite the fact that splice factors have been investigated only recently, they seem to play a prominent role in already five hallmarks of cancer: angiogenesis, resisting cell death, sustaining proliferation, deregulating cellular energetics and invasion and metastasis formation by affecting major signaling pathways such as epithelial-to-mesenchymal transition, the Warburg effect, DNA damage response and hormone receptor dependent proliferation. -

Stefano Sellitto 19-05-2020.Pdf
RNA Binding Proteins: from physiology to pathology An update Stefano Sellitto RBPs regulate the RNA metabolism A ‘conventional’ RNA-Binding Protein (RBP) participates in the formation of ribonucleoprotein (RNP) complexes that are principally involved in gene expression. Keene, 2007 High-Throughuput sequencing to study the RBPs world RIP-Seq (i)CLIP PAR-CLIP Hentze et al., 2018 The concept of RNA operon The RBPs profile is highly dynamic The combinatorial association of many RBPs acting in trans on RNA molecules results in the metabolic regulation of a distinct RNA subpopulations. Keene, 2007 The molecular features of protein-RNA interactions "Classical" RNA-Binding Domains Lunde et al., 2007 "Classical" RNA-Binding Domains RNA-recognition motif (RRM) double-stranded RNA-binding motif (dsRBM) Zinc-finger motif Stefl et al., 2005 Modularity of RBPs RBPs are usually composed by several repeated domains Lunde et al., 2007 Expanding the concept of the RNA Binding Hentze et al., 2018 Novel types of RNA binding Hentze et al., 2018 Lunde et al., 2007 RNA metabolism and neurological diseases RNA binding proteins preserves neuronal integrity De Conti et al., 2017 Alterations of RBPs in neurological disorders Nussbacher et al., 2019 Microsatellite expansion in FXS: reduced FMR1 transcription The reduction of FMRP levels induces an overexpressed LTD activity Bassell & Warren, 2008 Huber et al., 2002 Microsatellite expansion in FXTAS: FMR1 RNA-meidated toxicity FMR1 expanded RNA impairs the miRNA processing Sellier et al., 2013 Loss-of-function VS Gain-of-function Park et al., 2015 TDP-43 and FUS/TLS Ling et al., 2013 TDP-43 and FUS/TLS in ALS and FTD Ling et al., 2013 Cookson, 2017 Loss-of-function VS Gain-of-function Nucelar clearance (LOF) TDP-43 Nuclear TDP-43 (LOF) Loss of TDP-43 negative autoregulation Nucleus Abolishing RNA binding ability Cytoplasmatic stress (GOF) mitigates mutant TDP-43 toxicity Prevent mutant TDP-43 toxic activity on RNAs (GOF) Ihara et al., 2013 . -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

BTG2: a Rising Star of Tumor Suppressors (Review)
INTERNATIONAL JOURNAL OF ONCOLOGY 46: 459-464, 2015 BTG2: A rising star of tumor suppressors (Review) BIjING MAO1, ZHIMIN ZHANG1,2 and GE WANG1 1Cancer Center, Institute of Surgical Research, Daping Hospital, Third Military Medical University, Chongqing 400042; 2Department of Oncology, Wuhan General Hospital of Guangzhou Command, People's Liberation Army, Wuhan, Hubei 430070, P.R. China Received September 22, 2014; Accepted November 3, 2014 DOI: 10.3892/ijo.2014.2765 Abstract. B-cell translocation gene 2 (BTG2), the first 1. Discovery of BTG2 in TOB/BTG gene family gene identified in the BTG/TOB gene family, is involved in many biological activities in cancer cells acting as a tumor The TOB/BTG genes belong to the anti-proliferative gene suppressor. The BTG2 expression is downregulated in many family that includes six different genes in vertebrates: TOB1, human cancers. It is an instantaneous early response gene and TOB2, BTG1 BTG2/TIS21/PC3, BTG3 and BTG4 (Fig. 1). plays important roles in cell differentiation, proliferation, DNA The conserved domain of BTG N-terminal contains two damage repair, and apoptosis in cancer cells. Moreover, BTG2 regions, named box A and box B, which show a high level of is regulated by many factors involving different signal path- homology to the other domains (1-5). Box A has a major effect ways. However, the regulatory mechanism of BTG2 is largely on cell proliferation, while box B plays a role in combination unknown. Recently, the relationship between microRNAs and with many target molecules. Compared with other family BTG2 has attracted much attention. MicroRNA-21 (miR-21) members, BTG1 and BTG2 have an additional region named has been found to regulate BTG2 gene during carcinogenesis. -

Save Pdf (0.04
58 cambridge.org/jcts 2172 with these clinical observations, we observed altered myelopoiesis in HnrnpkTg mice. These mice demonstrate increased CD11b + Gr1 + populations in the Association between CYP450 polymorphisms and the bone marrow and peripheral blood. Indeed, these mice develop myeloid use of complementary medicine among patients with leukemia, indicated by >20% of circulating white blood cells harboring markers drug-resistant epilepsy in Puerto Rico of immature stem cells in conjunction with positive myeloperoxidase staining Bianca A. Torres-Hernández, Miriam E. Ríos Motta, Adrián Llerenaes and blast-appearing morphology. RPPA revealed expression of c-Myc positively correlated with increased hnRNP K levels. In HnrnpkTg mice, c-Myc protein and Jorge Duconge was increased, yet MYC RNA was invariably decreased compared to wildtype. University of Puerto Rico-Medical Sciences Campus, San Juan, Puerto To decipher a mechanism by which this may occur, we demonstrated a post- Rico transcriptional interaction between hnRNP K and c-Myc in vivo. JQ1, a BRD4 inhibitor, that epigenetically decreases c-Myc expression showed preferential activity against myeloid cells expressing high levels of hnRNP K both in vitro and OBJECTIVES/SPECIFIC AIMS: Patients with epilepsy often combine their in vivo. DISCUSSION/SIGNIFICANCE OF IMPACT: These preliminary studies antiepileptic drugs (AEDs) with complementary medicine (CM). They use CM demonstrate that hnRNP K overexpression causes myeloid malignancies in to treat their symptoms of comorbidities disorder, to reduce the side effect of both mouse and man. We have determined that c-Myc contributes in part to the AEDs or trying to achieve better control of their seizures. However, the hnRNP K-mediated leukemogenesis, and that targeting c-Myc may be an inconsistent patters of the use of CM among countries have been attributed to effective strategy for hnRNP K-overexpressing AML. -

Colon Cancer and Protein Arginine Methyltransferase 1 Gene Expression
ANTICANCER RESEARCH 29: 1361-1366 (2009) Colon Cancer and Protein Arginine Methyltransferase 1 Gene Expression ALEXANDRA PAPADOKOSTOPOULOU1*, KONSTANTINA MATHIOUDAKI2*, ANDREAS SCORILAS3, DIMITRIOS XYNOPOULOS1, ALEXANDROS ARDAVANIS4, ELIAS KOUROUMALIS5 and MAROULIO TALIERI2 Departments of 1Gastroenterology and 2Cellular Physiology, G. Papanicolaou Research Center of Oncology, and 4Oncology, St. Savvas Hospital, Athens; 3Department of Biochemistry and Molecular Biology, Faculty of Biology, University of Athens; 5Department of Gastroenterology, University Hospital of Heraklion, Crete, Greece Abstract. Background: In this study, the possible relation synthesis. Some of these modifications are reversible, such of the expression pattern of arginine methyltransferase 1 and as protein phosphorylation reactions, whereas others are colon cancer progression is investigated. Materials and apparently irreversible and can effectively create new types Methods: Colon cancer samples as well as normal colon of amino acids to broaden the chemical diversity of samples were used to define the arginine methyltransferase polypeptides. In this latter group of modifications, a 1 expression by RT-PCR. The results were associated with number of methylation reactions is included (1). Protein clinical and histological parameters of the tissues. Results: methylation involves transfer of a methyl group from S- In colon cancer tissue, only PRMT1 variants v1 and v2 were adenosylmethionine to acceptor groups on substrate often expressed. Statistical significance for the -

PRMT1, Human Recombinant Protein (Active) HMT2, HRMT1L2, IR1B4 Catalog # Pbv10454r
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 PRMT1, human recombinant protein (Active) HMT2, HRMT1L2, IR1B4 Catalog # PBV10454r Specification PRMT1, human recombinant protein PRMT1, human recombinant protein (Active) - (Active) - Background Product info PRMT1 methylate’s (mono & asymmetric Primary Accession Q99873 dimethylation) the guanidino nitrogens of Calculated MW 84.0 kDa KDa arginyl residues present in a glycine and arginine-rich domain (may methylate HNRNPA1 and histones) methylate’s SUPT5H. The PRMT1 PRMT1, human recombinant protein (Active) - Additional Info protein functions as a histone methyltransferase specific for H4. PRMT1 is an essential factor in oncogenesis and is a Gene ID 3276 potential novel therapeutic target in cancer. Gene Symbol ANM1 PRMT1-mediated methylation serves as a Other Names positive modulator of IR/IRS-1/PI3K pathway Protein arginine N-methyltransferase 1, and glucose uptake in skeletal muscle cells. Histone-arginine N-methyltransferase CAF1 is a new regulator of PRMT1-dependent PRMT1, Interferon receptor 1-bound protein 4, Histone-arginine N-methyltransferase arginine methylation. PRMT1 PRMT1, Interferon receptor 1-bound protein arginine-methylate’s MRE11 therefore it 4 regulates the activity of MRE11-RAD50-NBS1 complex during the intra-S-phase DNA damage Gene Source Human checkpoint response. PRMT1 plays a Source E. coli post-translationally part in regulating the Assay&Purity SDS-PAGE; ≥90% transcriptional activity. PRMT1 is found predominantly in the cytoplasm, though a Assay2&Purity2 HPLC; fraction of PRMT1 is located in the nucleus. Recombinant Yes PRMT1 Human Recombinant (a.a. 1-353) fused Sequence MHHHHHHMKI with His-MBP tag at N-terminus produced in EEGKLVIWIN E.Coli is a single, non-glycosylated, polypeptide GDKGYNGLAE chain containing 750 amino acids and having a VGKKFEKDTG molecular mass of 84 kDa. -

KH Domain Containing RNA-Binding Proteins Coordinate with Micrornas to Regulate
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.03.235127; this version posted August 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 KH domain containing RNA-binding proteins coordinate with microRNAs to regulate 2 Caenorhabditis elegans development. 3 4 Haskell D 1, Zinovyeva A1*. 5 6 7 (1) Division of Biology. Kansas State University. Manhattan, KS, 66506 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.03.235127; this version posted August 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 24 Running title: KH domain proteins coordinate with microRNAs, C. elegans 25 26 27 28 29 30 Keywords: microRNA, RNA binding protein, KH domain, hnRNPK 31 32 33 34 35 * Corresponding author: Anna Zinovyeva, PhD, 28 Ackert Hall, 1717 Claflin Road, Manhattan, 36 KS 66506, phone: 1-785-532-7727, email: [email protected] 37 38 39 40 41 42 43 44 45 46 2 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.03.235127; this version posted August 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Genetic and Pharmacological Approaches to Preventing Neurodegeneration
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2012 Genetic and Pharmacological Approaches to Preventing Neurodegeneration Marco Boccitto University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Neuroscience and Neurobiology Commons Recommended Citation Boccitto, Marco, "Genetic and Pharmacological Approaches to Preventing Neurodegeneration" (2012). Publicly Accessible Penn Dissertations. 494. https://repository.upenn.edu/edissertations/494 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/494 For more information, please contact [email protected]. Genetic and Pharmacological Approaches to Preventing Neurodegeneration Abstract The Insulin/Insulin-like Growth Factor 1 Signaling (IIS) pathway was first identified as a major modifier of aging in C.elegans. It has since become clear that the ability of this pathway to modify aging is phylogenetically conserved. Aging is a major risk factor for a variety of neurodegenerative diseases including the motor neuron disease, Amyotrophic Lateral Sclerosis (ALS). This raises the possibility that the IIS pathway might have therapeutic potential to modify the disease progression of ALS. In a C. elegans model of ALS we found that decreased IIS had a beneficial effect on ALS pathology in this model. This beneficial effect was dependent on activation of the transcription factor daf-16. To further validate IIS as a potential therapeutic target for treatment of ALS, manipulations of IIS in mammalian cells were investigated for neuroprotective activity. Genetic manipulations that increase the activity of the mammalian ortholog of daf-16, FOXO3, were found to be neuroprotective in a series of in vitro models of ALS toxicity. -
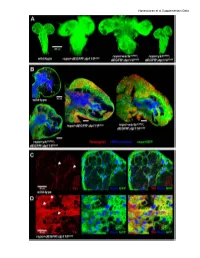
Supplementary Data Vigneswaran Et Al Supplementary Data
Vigneswaran et al Supplementary Data Vigneswaran et al Supplementary Data Figure S1: Yki is required for EGFR-PI3K-driven glial neoplasia in Drosophila (A) Optical projections of whole brain-nerve cord complexes from 3rd instar larvae approximately 130 hrs old. Dorsal view; anterior up. CD8-GFP (green) labels glial cell bodies. Compared to repo>dEGFRλ;dp110CAAX, warts knockdown (repo>wartsdsRNA; dEGFRλ;dp110CAAX) increased neoplastic brain overgrowth and yki knockdown (repo>ykidsRNA;dEGFRλ;dp110CAAX) decreased neoplastic brain overgrowth. (B) 3 µm optical projections of brain hemispheres, age-matched 3rd instar larvae. Frontal sections; anterior up; midline to left. Repo (red) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies; anti-HRP (blue) counter-stains for neurons and neuropil. (middle) repo>dEGFRλ;dp110CAAX showed increased glial cell numbers (red nuclei) compared to (upper left) wild-type. Compared to repo>dEGFRλ;dp110CAAX, (right) warts knockdown increased neoplastic glial cell numbers (red nuclei), whereas (lower left) yki knockdown reduced neoplastic glial cell numbers (red nuclei). (C, D) Low levels of Yki protein (red) was observed in wild-type central brain glia (white arrows, left panel in C) compared to high levels of cytoplasmic and nuclear Yki protein in dEGFRλ;dp110CAAX neoplastic glia (white arrows, left panel in D); Repo (blue) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies. Vigneswaran et al Supplementary Data Figure S2: YAP/TAZ expression confined to RTK-amplified tuMor cells and Maintained in patient-derived xenografts (A) On the left, immunohistochemical (IHC) staining in representative normal brain parenchyma in the cortex where YAP expression and TAZ expression was limited to vascular cells and was not detectable in normal neuronal and glial cells.