DNA-Binding and Transcriptional Activities of Human HSF4 Containing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Isyte: Integrated Systems Tool for Eye Gene Discovery
Lens iSyTE: Integrated Systems Tool for Eye Gene Discovery Salil A. Lachke,1,2,3,4 Joshua W. K. Ho,1,4,5 Gregory V. Kryukov,1,4,6 Daniel J. O’Connell,1 Anton Aboukhalil,1,7 Martha L. Bulyk,1,8,9 Peter J. Park,1,5,10 and Richard L. Maas1 PURPOSE. To facilitate the identification of genes associated ther investigation. Extension of this approach to other ocular with cataract and other ocular defects, the authors developed tissue components will facilitate eye disease gene discovery. and validated a computational tool termed iSyTE (integrated (Invest Ophthalmol Vis Sci. 2012;53:1617–1627) DOI: Systems Tool for Eye gene discovery; http://bioinformatics. 10.1167/iovs.11-8839 udel.edu/Research/iSyTE). iSyTE uses a mouse embryonic lens gene expression data set as a bioinformatics filter to select candidate genes from human or mouse genomic regions impli- ven with the advent of high-throughput sequencing, the cated in disease and to prioritize them for further mutational Ediscovery of genes associated with congenital birth defects and functional analyses. such as eye defects remains a challenge. We sought to develop METHODS. Microarray gene expression profiles were obtained a straightforward experimental approach that could facilitate for microdissected embryonic mouse lens at three key devel- the identification of candidate genes for developmental disor- opmental time points in the transition from the embryonic day ders, and, as proof-of-principle, we chose defects involving the (E)10.5 stage of lens placode invagination to E12.5 lens primary ocular lens. Opacification of the lens results in cataract, a leading cause of blindness that affects 77 million persons and fiber cell differentiation. -

A New Mutation in BFSP2 (G1091A) Causes Autosomal Dominant Congenital Lamellar Cataracts
Molecular Vision 2008; 14:1906-1911 <http://www.molvis.org/molvis/v14/a226> © 2008 Molecular Vision Received 17 August 2008 | Accepted 18 October 2008 | Published 24 October 2008 A new mutation in BFSP2 (G1091A) causes autosomal dominant congenital lamellar cataracts Xu Ma,1,2,3 Fei-Feng Li,1,2 Shu-Zhen Wang,4 Chang Gao,1,2 Meng Zhang,2 Si-Quan Zhu4 (The first three authors contributed equally to this work.) 1Graduate School, Peking Union Medical College, Beijing, China; 2Department of Genetics, National Research Institute for Family Planning, Beijing, China; 3WHO Collaborative Center for Research in Human Reproduction, Beijing, China; 4Beijing Tongren Eye Center, Capital Medical University, Beijing, China Purpose: We sought to identify the genetic defect in a four-generation Chinese family with autosomal dominant congenital lamellar cataracts and demonstrate the functional analysis with biosoftware of a candidate gene in the family. Methods: Family history data were recorded. Clinical and ophthalmologic examinations were performed on family members. All the members were genotyped with microsatellite markers at loci considered to be associated with cataracts. Two-point LOD scores were calculated by using the Linkage Software after genotyping. A mutation was detected by using gene-specific primers in direct sequencing. Wild type and mutant proteins were analyzed with Online Bio-Software. Results: Affected members of this family had lamellar cataracts. Linkage analysis was obtained at markers D3S2322 (LOD score [Z]=7.22, recombination fraction [θ]=0.0) and D3S1541 (Z=5.42, θ=0.0). Haplotype analysis indicated that the cataract gene was closely linked to these two markers. -

Identification of the Binding Partners for Hspb2 and Cryab Reveals
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2013-12-12 Identification of the Binding arP tners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non- Redundant Roles for Small Heat Shock Proteins Kelsey Murphey Langston Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Microbiology Commons BYU ScholarsArchive Citation Langston, Kelsey Murphey, "Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins" (2013). Theses and Dissertations. 3822. https://scholarsarchive.byu.edu/etd/3822 This Thesis is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston A thesis submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Master of Science Julianne H. Grose, Chair William R. McCleary Brian Poole Department of Microbiology and Molecular Biology Brigham Young University December 2013 Copyright © 2013 Kelsey Langston All Rights Reserved ABSTRACT Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactors and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston Department of Microbiology and Molecular Biology, BYU Master of Science Small Heat Shock Proteins (sHSP) are molecular chaperones that play protective roles in cell survival and have been shown to possess chaperone activity. -
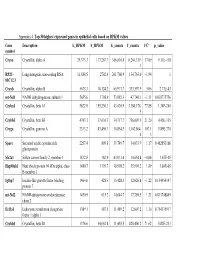
Appendix 4. Top 50 Highest Expressed Genes in Epithelial Cells Based on RPKM Values
Appendix 4. Top 50 highest expressed genes in epithelial cells based on RPKM values Gene Description E_RPKM F_RPKM E_counts F_counts FC* p_value symbol Cryaa Crystallin, alpha A 29,373.3 177,267.7 366,616.4 6,264,319. 17.09 9.11E-118 1 RP23– Long intergenic non-coding RNA 11,888.5 2702.4 261,760.9 134,763.0 −1.94 1 81C12.3 Cryab Crystallin, alpha B 5673.3 10,124.2 65,971.7 333,597.9 5.06 2.71E-43 mt-Nd1 NADH dehydrogenase, subunit 1 5655.6 1798.9 53,082.3 47,748.1 −1.11 0.838775756 Cryba1 Crystallin, beta A1 5622.0 155,230.3 43,420.9 3,380,176. 77.85 1.34E-240 5 Crybb3 Crystallin, beta B3 4743.1 37,636.3 34,717.7 736,007.9 21.20 4.45E-135 Cryga Crystallin, gamma A 2333.2 83,496.3 10,854.5 1,162,864. 107.1 5.89E-270 6 3 Sparc Secreted acidic cysteine rich 2257.4 809.8 39,749.7 34,033.9 −1.17 0.462853166 glycoprotein Slc2a1 Solute carrier family 2, member 1 1832.8 162.9 43,031.4 10,654.8 −4.04 1.67E-05 Hsp90ab1 Heat shock protein 90 kDa alpha, class 1480.7 1139.7 18,998.2 35,901.2 1.89 3.84E-05 B member 1 Igfbp7 Insulin-like growth factor binding 1464.6 428.3 15,428.3 12,626.8 −1.22 0.154954147 protein 7 mt-Nd2 NADH-ubiquinone oxidoreductase 1450.9 615.2 14,644.7 17,789.5 1.21 0.833748849 chain 2 Eef1a1 Eukaryotic translation elongation 1389.1 587.5 11,489.2 12,607.2 1.10 0.754135917 factor 1 alpha 1 Crybb1 Crystallin, beta B1 1376.6 34,662.8 11,455.5 820,406.2 71.62 5.82E-233 Htra3 HtrA serine peptidase 3 1338.6 162.0 23,197.6 6433.9 −3.61 3.93E-05 Gnb2l1 Guanine nucleotide-binding protein 1293.3 670.1 14,495.1 21,652.1 1.49 0.001685952 -

Localization of the Lens Intermediate Filament Switch by Imaging Mass Spectrometry
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.21.053793; this version posted April 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Localization of the Lens Intermediate Filament Switch by Imaging Mass Spectrometry Zhen Wang, Daniel J. Ryan, and Kevin L Schey* Department of Biochemistry, Vanderbilt University, Nashville, TN 37232 * To whom correspondence should be addressed Current address: Mass Spectrometry Research Center Vanderbilt University 465 21st Ave. So., Suite 9160 MRB III Nashville, TN 37232-8575 E-mail: [email protected] Phone: 615.936.6861 Fax: 615.343.8372 bioRxiv preprint doi: https://doi.org/10.1101/2020.04.21.053793; this version posted April 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Imaging mass spectrometry (IMS) enables targeted and untargeted visualization of the spatial localization of molecules in tissues with great specificity. The lens is a unique tissue that contains fiber cells corresponding to various stages of differentiation that are packed in a highly spatial order. The application of IMS to lens tissue localizes molecular features that are spatially related to the fiber cell organization. Such spatially resolved molecular information assists our understanding of lens structure and physiology; however, protein IMS studies are typically limited to abundant, soluble, low molecular weight proteins. In this study, a method was developed for imaging low solubility cytoskeletal proteins in the lens; a tissue that is filled with high concentrations of soluble crystallins. -

Variants in PAX6, PITX3 and HSF4 Causing Autosomal Dominant Congenital Cataracts ✉ ✉ Vanita Berry 1,2 , Alex Ionides2, Nikolas Pontikos 1,2, Anthony T
www.nature.com/eye ARTICLE OPEN Variants in PAX6, PITX3 and HSF4 causing autosomal dominant congenital cataracts ✉ ✉ Vanita Berry 1,2 , Alex Ionides2, Nikolas Pontikos 1,2, Anthony T. Moore2, Roy A. Quinlan3 and Michel Michaelides 1,2 © Crown 2021 BACKGROUND: Lens development is orchestrated by transcription factors. Disease-causing variants in transcription factors and their developmental target genes are associated with congenital cataracts and other eye anomalies. METHODS: Using whole exome sequencing, we identified disease-causing variants in two large British families and one isolated case with autosomal dominant congenital cataract. Bioinformatics analysis confirmed these disease-causing mutations as rare or novel variants, with a moderate to damaging pathogenicity score, with testing for segregation within the families using direct Sanger sequencing. RESULTS: Family A had a missense variant (c.184 G>A; p.V62M) in PAX6 and affected individuals presented with nuclear cataract. Family B had a frameshift variant (c.470–477dup; p.A160R*) in PITX3 that was also associated with nuclear cataract. A recurrent missense variant in HSF4 (c.341 T>C; p.L114P) was associated with congenital cataract in a single isolated case. CONCLUSIONS: We have therefore identified novel variants in PAX6 and PITX3 that cause autosomal dominant congenital cataract. Eye; https://doi.org/10.1038/s41433-021-01711-x INTRODUCTION consistent with early developmental effects as would be Cataract the opacification of the eye lens is the most common, but anticipated for PAX6 and PITX3 transcription factors. Recently, treatable cause of blindness in the world (https://www.who.int/ we have found two novel mutations in the transcription factors publications-detail/world-report-on-vision). -

A Comprehensive Analysis of the Expression of Crystallins in Mouse Retina Jinghua Xi Washington University School of Medicine in St
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2003 A comprehensive analysis of the expression of crystallins in mouse retina Jinghua Xi Washington University School of Medicine in St. Louis Rafal Farjo University of Michigan - Ann Arbor Shigeo Yoshida University of Michigan - Ann Arbor Timothy S. Kern Case Western Reserve University Anand Swaroop University of Michigan - Ann Arbor See next page for additional authors Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Recommended Citation Xi, Jinghua; Farjo, Rafal; Yoshida, Shigeo; Kern, Timothy S.; Swaroop, Anand; and Andley, Usha P., ,"A comprehensive analysis of the expression of crystallins in mouse retina." Molecular Vision.9,. 410-419. (2003). https://digitalcommons.wustl.edu/open_access_pubs/1801 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Jinghua Xi, Rafal Farjo, Shigeo Yoshida, Timothy S. Kern, Anand Swaroop, and Usha P. Andley This open access publication is available at Digital Commons@Becker: https://digitalcommons.wustl.edu/open_access_pubs/1801 Molecular Vision 2003; 9:410-9 <http://www.molvis.org/molvis/v9/a53> © 2003 Molecular Vision Received 28 May 2003 | Accepted 19 August 2003 | Published 28 August 2003 A comprehensive analysis of the expression of crystallins in mouse retina Jinghua Xi,1 Rafal Farjo,3 Shigeo Yoshida,3 Timothy S. Kern,5 Anand Swaroop,3,4 Usha P. Andley1,2 Departments of 1Ophthalmology and Visual Sciences and 2Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. -

Related Macular Degeneration and Cutis Laxa
UvA-DARE (Digital Academic Repository) Genetic studies of age-related macular degeneration Baas, D.C. Publication date 2012 Document Version Final published version Link to publication Citation for published version (APA): Baas, D. C. (2012). Genetic studies of age-related macular degeneration. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:05 Oct 2021 G������ S������ �� A��-������� M������ D����������� D����������� M������ G������ S������ �� A��-������� | 2012 D�������� C. B��� G������ S������ �� A��-������� M������ D����������� D�������� C. B��� cover.indd 1 31-10-12 08:36 Genetic Studies of Age-related Macular Degeneration Dominique C. Baas Chapter 0.indd 1 23-10-12 19:24 The research described in this thesis was conducted at the Netherlands Institute for Neuroscience (NIN), an institute of the Royal Netherlands Academy of Arts and Sciences, Department of Clinical and Molecular Ophthalmogenetics, Amsterdam, The Netherlands. -

Method for Diagnosing Non-Small Cell Lung Cancers
(19) TZZ Z _ T (11) EP 2 270 221 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 05.01.2011 Bulletin 2011/01 C12Q 1/68 (2006.01) C12N 15/12 (2006.01) G01N 33/50 (2006.01) C12N 15/11 (2006.01) (2006.01) (2006.01) (21) Application number: 10010329.0 A61P 35/00 A61K 39/00 (22) Date of filing: 22.09.2003 (84) Designated Contracting States: (72) Inventors: AT BE BG CH CY CZ DE DK EE ES FI FR GB GR • Nakamura, Yusuke HU IE IT LI LU MC NL PT RO SE SI SK TR Yokohama-shi Kanagawa 225-0011 (JP) (30) Priority: 30.09.2002 US 414673 P • Daigo, Yataro 28.02.2003 US 451374 P Yokohama-shi 28.04.2003 US 466100 P Kanagawa 222-0031 (JP) • Nakatsuru, Shuichi (62) Document number(s) of the earlier application(s) in Saitama-shi accordance with Art. 76 EPC: Saitama 338-0002 (JP) 06022167.8 / 1 743 947 03753941.8 / 1 551 998 (74) Representative: Vossius & Partner Siebertstrasse 4 (71) Applicant: Oncotherapy Science, Inc. 81675 München (DE) Kawasaki-shi Kanagawa 213-0012 (JP) Remarks: This application was filed on 23-09-2010 as a divisional application to the application mentioned under INID code 62. (54) Method for diagnosing non-small cell lung cancers (57) Disclosed are methods for detecting non-small cancerous tissues are provided. Also disclosed are meth- cell lung cancer using differentially expressed genes. ods of identifying compounds for treating and preventing Furthermore, novel human genes whose expression is non-small cell lung cancer. -

Common Variants in SOX-2 and Congenital Cataract Genes Contribute to Age-Related Nuclear Cataract
ARTICLE https://doi.org/10.1038/s42003-020-01421-2 OPEN Common variants in SOX-2 and congenital cataract genes contribute to age-related nuclear cataract Ekaterina Yonova-Doing et al.# 1234567890():,; Nuclear cataract is the most common type of age-related cataract and a leading cause of blindness worldwide. Age-related nuclear cataract is heritable (h2 = 0.48), but little is known about specific genetic factors underlying this condition. Here we report findings from the largest to date multi-ethnic meta-analysis of genome-wide association studies (discovery cohort N = 14,151 and replication N = 5299) of the International Cataract Genetics Consortium. We confirmed the known genetic association of CRYAA (rs7278468, P = 2.8 × 10−16) with nuclear cataract and identified five new loci associated with this dis- ease: SOX2-OT (rs9842371, P = 1.7 × 10−19), TMPRSS5 (rs4936279, P = 2.5 × 10−10), LINC01412 (rs16823886, P = 1.3 × 10−9), GLTSCR1 (rs1005911, P = 9.8 × 10−9), and COMMD1 (rs62149908, P = 1.2 × 10−8). The results suggest a strong link of age-related nuclear cat- aract with congenital cataract and eye development genes, and the importance of common genetic variants in maintaining crystalline lens integrity in the aging eye. #A list of authors and their affiliations appears at the end of the paper. COMMUNICATIONS BIOLOGY | (2020) 3:755 | https://doi.org/10.1038/s42003-020-01421-2 | www.nature.com/commsbio 1 ARTICLE COMMUNICATIONS BIOLOGY | https://doi.org/10.1038/s42003-020-01421-2 ge-related cataract is the leading cause of blindness, structure (meta-analysis genomic inflation factor λ = 1.009, accounting for more than one-third of blindness Supplementary Table 4 and Supplementary Fig. -

Congenital Cataracts Due to a Novel 2‑Bp Deletion in CRYBA1/A3
1614 MOLECULAR MEDICINE REPORTS 10: 1614-1618, 2014 Congenital cataracts due to a novel 2‑bp deletion in CRYBA1/A3 JING ZHANG1, YANHUA ZHANG1, FANG FANG1, WEIHONG MU1, NING ZHANG2, TONGSHUN XU3 and QINYING CAO1 1Prenatal Diagnosis Center, Shijiazhuang Obstetrics and Gynecology Hospital; 2Department of Cardiology, The Second Hospital of Hebei Medical University; 3Department of Surgery, Shijiazhuang Obstetrics and Gynecology Hospital, Shijiazhuang, Hebei, P.R. China Received September 22, 2013; Accepted April 11, 2014 DOI: 10.3892/mmr.2014.2324 Abstract. Congenital cataracts, which are a clinically and located in the eye lens. The major human crystallins comprise genetically heterogeneous group of eye disorders, lead to 90% of protein in the mature lens and contain two different visual impairment and are a significant cause of blindness superfamilies: the small heat‑shock proteins (α-crystallins) in childhood. A major proportion of the causative mutations and the βγ-crystallins. for congenital cataracts are found in crystallin genes. In the In this study a functional candidate approach was used present study, a novel deletion mutation (c.590-591delAG) in to investigate the known crystallin genes, including CRYAA, exon 6 of CRYBA1/A3 was identified in a large family with CRYAB, CRYBA1/A3, CRYBB1, CRYBB2, CRYGC, CRYGD autosomal dominant congenital cataracts. An increase in and CRYGS, in which a major proportion of the mutations local hydrophobicity was predicted around the mutation site; identified in a large family with congenital cataracts were however, further studies are required to determine the exact found. effect of the mutation on βA1/A3-crystallin structure and function. To the best of our knowledge, this is the first report Subjects and methods of an association between a frameshift mutation in exon 6 of CRYBA1/A3 and congenital cataracts. -

Proteome Profiling of Developing Murine Lens Through Mass
Lens Proteome Profiling of Developing Murine Lens Through Mass Spectrometry Shahid Y. Khan,1 Muhammad Ali,1 Firoz Kabir,1 Santosh Renuse,2 Chan Hyun Na,2 C. Conover Talbot Jr,3 Sean F. Hackett,1 and S. Amer Riazuddin1 1The Wilmer Eye Institute, Johns Hopkins University School of Medicine, Baltimore, Maryland, United States 2Department of Biological Chemistry, Johns Hopkins University School of Medicine, Baltimore, Maryland, United States 3Institute for Basic Biomedical Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, United States Correspondence: S. Amer Riazuddin, PURPOSE. We previously completed a comprehensive profile of the mouse lens transcriptome. The Wilmer Eye Institute, Johns Here, we investigate the proteome of the mouse lens through mass spectrometry–based Hopkins University School of Medi- protein sequencing at the same embryonic and postnatal time points. cine, 600 N. Wolfe Street; Maumenee 840, Baltimore, MD 21287, USA; METHODS. We extracted mouse lenses at embryonic day 15 (E15) and 18 (E18) and postnatal [email protected]. day 0 (P0), 3 (P3), 6 (P6), and 9 (P9). The lenses from each time point were preserved in three Submitted: February 2, 2017 distinct pools to serve as biological replicates for each developmental stage. The total cellular Accepted: October 13, 2017 protein was extracted from the lens, digested with trypsin, and labeled with isobaric tandem mass tags (TMT) for three independent TMT experiments. Citation: Khan SY, Ali M, Kabir F, et al. Proteome profiling of developing mu- RESULTS. A total of 5404 proteins were identified in the mouse ocular lens in at least one rine lens through mass spectrometry.