Practical Breeding of Red-Fleshed Apple: Cultivar Combination For
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Apples Catalogue 2019
ADAMS PEARMAIN Herefordshire, England 1862 Oct 15 Nov Mar 14 Adams Pearmain is a an old-fashioned late dessert apple, one of the most popular varieties in Victorian England. It has an attractive 'pearmain' shape. This is a fairly dry apple - which is perhaps not regarded as a desirable attribute today. In spite of this it is actually a very enjoyable apple, with a rich aromatic flavour which in apple terms is usually described as Although it had 'shelf appeal' for the Victorian housewife, its autumnal colouring is probably too subdued to compete with the bright young things of the modern supermarket shelves. Perhaps this is part of its appeal; it recalls a bygone era where subtlety of flavour was appreciated - a lovely apple to savour in front of an open fire on a cold winter's day. Tree hardy. Does will in all soils, even clay. AERLIE RED FLESH (Hidden Rose, Mountain Rose) California 1930’s 19 20 20 Cook Oct 20 15 An amazing red fleshed apple, discovered in Aerlie, Oregon, which may be the best of all red fleshed varieties and indeed would be an outstandingly delicious apple no matter what color the flesh is. A choice seedling, Aerlie Red Flesh has a beautiful yellow skin with pale whitish dots, but it is inside that it excels. Deep rose red flesh, juicy, crisp, hard, sugary and richly flavored, ripening late (October) and keeping throughout the winter. The late Conrad Gemmer, an astute observer of apples with 500 varieties in his collection, rated Hidden Rose an outstanding variety of top quality. -

Variety Description Origin Approximate Ripening Uses
Approximate Variety Description Origin Ripening Uses Yellow Transparent Tart, crisp Imported from Russia by USDA in 1870s Early July All-purpose Lodi Tart, somewhat firm New York, Early 1900s. Montgomery x Transparent. Early July Baking, sauce Pristine Sweet-tart PRI (Purdue Rutgers Illinois) release, 1994. Mid-late July All-purpose Dandee Red Sweet-tart, semi-tender New Ohio variety. An improved PaulaRed type. Early August Eating, cooking Redfree Mildly tart and crunchy PRI release, 1981. Early-mid August Eating Sansa Sweet, crunchy, juicy Japan, 1988. Akane x Gala. Mid August Eating Ginger Gold G. Delicious type, tangier G Delicious seedling found in Virginia, late 1960s. Mid August All-purpose Zestar! Sweet-tart, crunchy, juicy U Minn, 1999. State Fair x MN 1691. Mid August Eating, cooking St Edmund's Pippin Juicy, crisp, rich flavor From Bury St Edmunds, 1870. Mid August Eating, cider Chenango Strawberry Mildly tart, berry flavors 1850s, Chenango County, NY Mid August Eating, cooking Summer Rambo Juicy, tart, aromatic 16th century, Rambure, France. Mid-late August Eating, sauce Honeycrisp Sweet, very crunchy, juicy U Minn, 1991. Unknown parentage. Late Aug.-early Sept. Eating Burgundy Tart, crisp 1974, from NY state Late Aug.-early Sept. All-purpose Blondee Sweet, crunchy, juicy New Ohio apple. Related to Gala. Late Aug.-early Sept. Eating Gala Sweet, crisp New Zealand, 1934. Golden Delicious x Cox Orange. Late Aug.-early Sept. Eating Swiss Gourmet Sweet-tart, juicy Switzerland. Golden x Idared. Late Aug.-early Sept. All-purpose Golden Supreme Sweet, Golden Delcious type Idaho, 1960. Golden Delicious seedling Early September Eating, cooking Pink Pearl Sweet-tart, bright pink flesh California, 1944, developed from Surprise Early September All-purpose Autumn Crisp Juicy, slow to brown Golden Delicious x Monroe. -

An Old Rose: the Apple
This is a republication of an article which first appeared in the March/April 2002 issue of Garden Compass Magazine New apple varieties never quite Rosaceae, the rose family, is vast, complex and downright confusing at times. completely overshadow the old ones because, as with roses, a variety is new only until the next This complexity has no better exemplar than the prince of the rose family, Malus, better known as the variety comes along and takes its apple. The apple is older in cultivation than the rose. It presents all the extremes in color, size, fragrance place. and plant character of its rose cousin plus an important added benefit—flavor! One can find apples to suit nearly every taste and cultural demand. Without any special care, apples grow where no roses dare. Hardy varieties like the Pippins, Pearmains, Snow, Lady and Northern Spy have been grown successfully in many different climates across the U.S. With 8,000-plus varieties worldwide and with new ones introduced annually, apple collectors in most climates are like kids in a candy store. New, Favorite and Powerhouse Apples New introductions such as Honeycrisp, Cameo and Pink Lady are adapted to a wide range of climates and are beginning to be planted in large quantities. The rich flavors of old favorites like Spitzenburg and Golden Russet Each one is a unique eating experience that are always a pleasant surprise for satisfies a modern taste—crunchy firmness, plenty inexperienced tasters. of sweetness and tantalizing flavor. Old and antique apples distinguish These new varieties show promise in the themselves with unusual skin competition for the #1 spot in the world’s colors and lingering aftertastes produce sections and farmers’ markets. -

Cold Damage Cultivar Akero 0 Albion 0 Alexander 0 Alkmene 0 Almata 0
Cold Damage Table 16 1. less than 5% Bud 118 0 2. 5-15% Bud 9 on Ranetka 0 3. More than 15%. Cultivar 4. severe (50% ) Carroll 0 Akero 0 Centennial 0 Albion 0 Chehalis 0 Alexander 0 Chestnut Crab 0 Alkmene 0 Collet 0 Almata 0 Collins 0 American Beauty 0 Crab 24 false yarlington 0 Anaros 0 Cranberry 0 Anoka 0 Croncels 0 Antonovka 81 0 Dan Silver 0 Antonovka 102 0 Davey 0 Antonovka 109 0 Dawn 0 antonovka 52 0 Deane 0 Antonovka 114 0 Dolgo (grafted) 0 Antonovka 1.5 0 Douce Charleviox 0 Antonovka 172670-B 0 Duchess 0 Antonovka 37 0 Dudley 0 Antonovka 48 0 Dudley Winter 0 Antonovka 49 0 Dunning 0 Antonovka 54 0 Early Harvest 0 Antonovka Debnicka 0 Elstar 0 Antonovka Kamenichka 0 Equinox 0 Antonovka Monasir 0 Erwin Bauer 0 Antonovka Shafrain 0 Fameuse 0 Aroma 0 Fantazja 0 Ashmead's Kernal 0 Fox Hill 0 Audrey 0 Frostbite TM 0 Autumn Arctic 0 Garland 0 Baccata 0 Geneva 0 Banane Amere 0 Gideon 0 Beacon 0 Gilpin 0 Beautiful Arcade 0 Gingergold 0 Bedford 0 Golden Russet 0 Bessemianka Michurina 0 Granny Smith Seedling 0 Bilodeau 0 Green Peak 0 Black Oxford 0 Greenkpeak 0 Blue Pearmain 0 Greensleeves 0 Borovitsky 0 Haralred 0 Breaky 0 Haralson 0 Cold Damage Table 16 1. less than 5% McIntosh 0 2. 5-15% Melba 0 3. More than 15%. Cultivar 4. severe (50% ) Miami 0 Harcourt 0 Minnehaha 0 Hawaii 0 MN 85-22-99 0 Herring's Pippin 0 MN 85-23-21 0 Hewe's Crab 0 MN 85-27-43 0 Hiburnal 0 Morden 0 Honeygold 0 Morden 359 0 Hyslop Crab 0 Niedzwetzkyana 0 Island Winter 0 No Blow 0 Jersey Mac 0 Noran 0 Jonamac 0 Noret 0 Jonathan 0 Norhey 0 Kazakh 1 0 Norland 0 Kazakh -

Brightonwoods Orchard
Managing Diversity Jimmy Thelen Orchard Manager at Brightonwoods Orchard 2020 Practical Farmers of Iowa Presentation MAP ORCHARD PEOPLE ORCHARD PEOPLE • UW-Parkside Graduate • Started at Brightonwoods in 2006 • Orchard Manager and in charge of Cider House • Case Tractor Hobby & Old Abe's News ORCHARD HISTORY • Initial sales all from on the farm (1950- 2001) “Hobby Orchard” • Expansion into multiple cultivars (10 acres) • 1980's • Added refrigeration • Sales building constructed ORCHARD HISTORY • Retirement begets new horizons • (1997-2020) • Winery (2000-2003) additional 2 acres of trees for the winery and 30+ varieties of apples & pears ORCHARD HISTORY • Cider House (2006) with UV light treatment and contract pressing • Additional ½ acre of Honeycrisp ORCHARD HISTORY • Additional 3 acres mixed variety higher density planting ~600 trees per acre ORCHARD HISTORY • Addition of 1 acre of River Belle and Pazazz ORCHARD • Not a Pick- your-own • All prepicked and sorted • Not Agri- entertainment focused ACTIVITIES WHERE WE SELL • Retail Focused • At the Orchard • Summer / Fall Farmers' Markets • Winter Farmers' Markets • Restaurants • Special Events ADDITIONAL PRODUCTS • Honey, jams & jellies • Pumpkins & Gourds • Squash & Garlic • Organic vegetables on Sundays • Winery Products • Weekend snacks and lunches 200+ VARIETIES Hubardtson Nonesuch (October) Rambo (September) Americus Crab (July / August) Ida Red (October) Red Astrashan (July–August) Arkansas Black (October) Jersey Mac (July–August) Red Cortland(September) Ashmead's Kernal (October) -

Australian Fruitgrower APAL’S CEO Report
YOUR LINK TO INNOVATION CONNECT WITH INDUSTRY, COLLABORATE AND CULTIVATE IDEAS. BECOME A MEMBER NOW. MEMBERSHIP IS FREE AND IT ONLY TAKES A FEW MINUTES TO APPLY ONLINE AT WWW.HORTICULTURE.COM.AU /MEMBERSHIP. CALL 1300 880 981 FOR MORE INFORMATION. CONTENTS Australian Fruitgrower APAL’s CEO report . 4 Publishers APAL news . 5 Apple and Pear Australia Limited (APAL) is a not-for-profit organisation that supports and provides services to Australia’s commercial apple and pear growers. EVENTS Suite G01, 128 Jolimont Road, Fruit Logistica . .8 East Melbourne VIC 3002 t: (03) 9329 3511 f: (03) 9329 3522 Pink Lady ® exporters’ meeting . .9 w: www.apal.org.au 08 Managing Editor State Roundup . 10 Currie Communications e: [email protected] Technical Editor EXPORT & MARKETING Angus Crawford e: [email protected] A ripe time to export . .12 Hort Innovation marketing update . .15 Online Manager Richelle Zealley PROFILE e: [email protected] Flying high with Lenswood . .18 Advertising The publishers accept no responsibility for the contents of advertisements. All advertisements are 18 accepted in good faith and the liability of advertising content is the responsibility of the advertiser. ORCHARD MANAGEMENT Hyde Media e: [email protected] Labour-saving harvesting . .22 Graphic Design Pome fruit rootstocks . .26 Vale Graphics e: [email protected] POST-HARVEST Post-harvest seminar wrap . .30 Copyright All material in Australian Fruitgrower is copyright. NO material can be reproduced in whole or in part without the permission of the publisher. R&D update . 34 While every effort is made to ensure the accuracy of Weather, quiz and crossword . -

Assessment of One Year of Growth in the New Jersey Hard Cider Variety Trial M
Assessment of One Year of Growth in the New Jersey Hard Cider Variety Trial M. Muehlbauer and R. Magron Rutgers University There is much interest in hard cider in New Jersey. the best apples for their cider. Some traditional fresh In New Jersey the manufacture of hard cider is covered market apples make good hard cider, but many of the under the Farm Winery Act, passed in 1981. NJ law hard cider producers are looking for both the English treats hard cider as a type of wine as it is fermented and French hard cider varieties to source for production from fruits (N.J.A.C. 18:3-1.2) of craft hard ciders. As such there is much interest from existing sweet Apple growers and hard cider producers are look- cider producers to make and sell hard cider as a value ing to source these hard cider apple varieties that have added product. There is also great interest and for the specifi c characteristics for craft hard cider. There is an establishment of new, stand alone hard cideries. NJ now abundant interest and momentum from these NJ hard has a mix of both established, seen the list at https:// cider producers to evaluate and grow or purchase these www.ciderculture.com/cideries/state/nj/ varieties from other apple growers. These hard cider producers all need a supply of As a result, it is important to establish a demonstra- ϴϬ ϳϬ ϲϬ ϱϬ ϰϬ ϯϬ ϮϬ $YHUDJH +HLJKW LQ $YHUDJH 'LDPHWHU PP ϭϬ Ϭ /RGL 0DMRU /LQGHO 0DUJLO &ROODRV 'DELQHWW +DUULVRQ :LFNVRQ )R[ZKHOS 0DULDOHQD %ODQTXLQD (OOLV%LWWHU 3LQN3HDUO 6WRNH5HG 3LHOGH6DSD &DOYLOOH%ODQF %OXH3HDUPDLQ -

Genetic Diversity of Red-Fleshed Apples (Malus)
Euphytica DOI 10.1007/s10681-011-0579-7 Genetic diversity of red-fleshed apples (Malus) Steven van Nocker • Garrett Berry • James Najdowski • Roberto Michelutti • Margie Luffman • Philip Forsline • Nihad Alsmairat • Randy Beaudry • Muraleedharan G. Nair • Matthew Ordidge Received: 31 May 2011 / Accepted: 3 November 2011 Ó Springer Science+Business Media B.V. 2011 Abstract Anthocyanins are flavonoid pigments including the core and cortex (flesh). Red-fleshed imparting red, blue, or purple pigmentation to fruits, apple genotypes are an attractive starting point for flowers and foliage. These compounds are powerful development of novel varieties for consumption and antioxidants in vitro, and are widely believed to nutraceutical use through traditional breeding and contribute to human health. The fruit of the domestic biotechnology. However, cultivar development is apple (Malus x domestica) is a popular and important limited by lack of characterization of the diversity source of nutrients, and is considered one of the top of genetic backgrounds showing this trait. We iden- ‘functional foods’—those foods that have inherent tified and cataloged red-fleshed apple genotypes from health-promoting benefits beyond basic nutritional four Malus diversity collections representing over value. The pigmentation of typical red apple fruits 3,000 accessions including domestic cultivars, wild results from accumulation of anthocyanin in the skin. species, and named hybrids. We found a striking However, numerous genotypes of Malus are known range of flesh color intensity -

EFM-2017-05 EN Article Guerra
Walter Guerra Laimburg Research Centre [email protected] The hunt for new future apple varieties Info In the course of last year, various announcements appeared in the media that symbolised the revival of variety innovation seen recently in the apple sector. Because the apple sec- Original article tor is currently in a crisis, innovations, which are exclusive to a greater or lesser degree, are The original article being incorporated in strategies to distinguish individual players from the competition appeared in Frutticol- (Sansavini and Guerra, 2015). tura 11 (2016), it was translated and amen- Three of the announcements mentioned above the more than 80 apple breeding programmes ded by Julia Strobl. were: in the world. The Breeding Group Midwest Apple Improvement Association (MAIA) has signed an agreement with the International Pome Fruit Alliance (IPA) for the Financing of variety breeding market introduction of the rst hybrids of MAIA under the brand name EverCrisp® (www.freshplaza. Many public institutes are spending less time com, 8/2/16). and money on the independent testing of new varieties. Moreover, numerous breed- „We have acquired the exclusive rights to three new ing programmes are no longer government varieties: Kizuri, Gradisca and Lumaga Galant®. The "nanced, but operate as semi-public organi- latter is resistant to scab“, announced the chair of sations or are even privatised. These pro- the Melinda consortium (Italiafruit News, 6/9/16). grammes are under enormous pressure to “We are currently working on new varieties that become pro"table in the short term. Con- will bring about a revolution in the apple sector“, tracts for the propagation, cultivation and/ explained the General Director of Apofruit Bastoni. -

Tree Physical Characteristics 1 Variety Vigor 36-305 Adam's Pearmain
Tree Physical Characteristics 1-few cankers 2- moderate (less than 10 lesions), non lethal 3- girdling, lethal or g-50-90% 1 - less than 5% extensive a- f-35-50% 2 - 5-15% anthracnose canker p -less than 35% 3 - More than 15% 3-strongest 1 yes or no n- nectria (from vertical) 4 - severe (50% ) alive or dead 1-weakest branch dieback/cold 1 Variety leaf scab canker angle damage mortality notes vigor 36-305 y a 1 g 1 a perfect form 3 1 weak, 1 decent, spindly Adam's Pearmain n a 1 p 2 a growth 2 Adanac n 0 f 3 a weak 1 no crop in 11 Akero y f a years 2 Albion n 0 f 0 a 2 only 10% of fruit had scab, none on leaves, Alexander n 0 f a bullseye on fruit 3 bad canker, Alkmene y a 1,3 f 0 a bushy 2 vigorous but Almata y f 0 a lanky 3 red 3/4 fruits, nice open form, Almey y 0 g 1 a severe scab 3 Altaiski Sweet y a Alton y g 1 a 3 American Beauty y f 0 a slow to leaf out 3 Amsib Crab n f 1 a 2 looks decent, Amur Deburthecort y 2 a cold damage early leaf drop, Anaros y 2 g 0 a nicely branched 3 dieback spurs Anis n a 1 , other cankers p 1 a only 1 Anise Reinette y a 1 f 2 a 2 Anisim y a 1 p 1 a leader dieback 3 Ann's n 0 f 2 a local variety 2 Anoka y f a 2 Antonovka 81 n g a 3 Antonovka 102 n g a 3 Antonovka 109 n f a 3 antonovka 52 n g a 3 Antonovka 114 n g a 3 Antonovka 1.5 n a1 g 0 a 3 n f 0 a 3 Antonovka 172670-B Page 1 of 17 Tree Physical Characteristics Antonovka 37 n g a 3 Antonovka 48 n g a 3 Antonovka 49 n g a 3 Antonovka 54 n f a slow to leaf out 3 Antonovka Debnicka n f 0 a 3 Antonovka Kamenichka n a 1 f 0 a 3 Antonovka Michurin n 0 f -
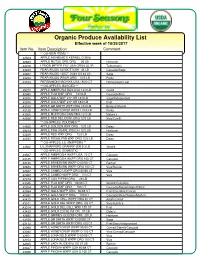
Organic Produce Availability List
$PLPRESS=FCPRT01; 10/26/17 Four Seasons Produce Co. PAGE Phone #: 800-422-8384 Fax #: (717) 721-2597 PRICE LIST P92002 FOUROrganic SEASONS PRODUCE/ORGANICSProduce Availability Attention: List 10/26/17 to 10/26/17 Effective week of 10/29/2017 Item#Item No.Item DescriptionItem Description Price Comment Comment ===== ==============================** OG-NEW ITEMS ** ======= ============================== 42884 ** OG-NEWAPPLE ITEMS ASHMEAD’S ** KERNEL O 28 lb 4288442883 APPLE ASHMEAD’SAPPLE MUTSU KERNEL ORE ORG O 28 28 lb LB 69.50 Heirloom 4288344016 APPLE MUTSULEMON MEYERORE ORG FCY USA ORG928 LB 20 LB 69.50 Tomorrow’s Heirloom 4401643243 LEMON MEYERPEAR ANJOU FCY USA80/90CT ORG9 US#1 20 44 LB LB 40.50 Cascade/Sage Tomorrow’s 4324343267 PEAR ANJOUPEAR ANJOU80/90CT 100CT US#1 US#1 44US 44LB LB 69.50 Sage Cascade/Sage 4326743239 PEAR ANJOUPEAR ANJOU100CT WASHUS#1 ORGUS 44 12/3 LB LB 65.50 Purity Sage 4323943189 PEAR ANJOUPERSIMMON WASH HACHIYAORG USA12/3 18/22 LB CT 39.00 Homegrown/Twin Purity 43189 PERSIMMON** OG-APPLES, HACHIYA BAGGEDUSA 18/22 ** CT 33.00 Homegrown/Twin 49220 ** OG-APPLES,APPLE AMBROSIA BAGGED BAG ** USA 12/3 LB Covilli 4922042995 APPLE AMBROSIAAPPLE FUJI BAGWXF ORGUSA 12/3 12/3 LBLB 48.00 Cascade/First Covilli 4299542975 APPLE FUJIAPPLE WXF GALA ORG WXF 2.5" OR12/3 12/3 LB LB 77.00 Sage/Independent Cascade/First 4297542978 APPLE GALAAPPLE WXF GALA 2.5" WXF 2.5"OR OR12/3 18/2 LB LB 42.50 First Sage/Independent 4297843100 APPLE GALAAPPLE WXF GR SMITH2.5" WXFOR ORG18/2 12/3 LB LB 45.50 Bridges/Stemilt First 4310043126 APPLE -

Heirloom Apple Varieties Better Sliced
om Orc eirlo hard H s Tr y these apples sliced with cheese. All apples taste Heirloom Apple Varieties better sliced. More delicate flesh and Our Apples Make History! less skin assures Late September Early October full apple flavor in every bite. Gravenstein: The Gravenstein is considered to be one of the best Grimes Golden: If you are a Golden Delicious fan, try Ho on all-around apples for baking, cooking and eating. it has a sweet, the parent, Grimes Golden. A clear, deep yellow skin od River, Oreg tart flavor and juicy, crisp texture. the Gravenstein is native to covers a fine grained, spicy flesh. Very juicy and excellent Denmark, discovered in 1699. It traveled to America with for cider. Its tender flesh keeps it from holding up well for Russian fur traders, who planted orchards at Fort Ross, CA in the baking. The Grimes Golden’s exceptional flavor keeps it a early 1800's. favorite dessert apple of many. Discovered in Brook County, Virginia in 1804 by Thomas Grimes. Pink Pearl: Cut or bite into this apple and you are in for a Certified Organic surprise. In fact, it is an offspring of another variety called Calville Blanc: This world renowned dessert apple dates 'Surprise'! Pink fleshed, pearly skinned, good tasting with sweet from 16th century France. Its’ flattened round shape makes to tart flavor. Makes pink applesauce and pretty fruit tarts. it distinctive looking, so much that Monet put it in his 1879 Aromatic flavor with a hint of grapefruit. painting “Apples and Grapes”. It has a tart, effervescent Late October flavor, and is good for eating.