Mitochondrial Genomics of Gadine Fishes: Implications for Taxonomy and Biogeographic Origins from Whole-Genome Data Sets
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Systematics of Gadid Fishes: Implications for the Biogeographic Origins of Pacific Species
Color profile: Disabled Composite Default screen 19 Molecular systematics of gadid fishes: implications for the biogeographic origins of Pacific species Steven M. Carr, David S. Kivlichan, Pierre Pepin, and Dorothy C. Crutcher Abstract: Phylogenetic relationships among 14 species of gadid fishes were investigated with portions of two mitochondrial DNA (mtDNA) genes, a 401 base pair (bp) segment of the cytochrome b gene, and a 495 bp segment of the cytochrome oxidase I gene. The molecular data indicate that the three species of gadids endemic to the Pacific Basin represent simultaneous invasions by separate phylogenetic lineages. The Alaskan or walleye pollock (Theragra chalcogramma) is about as closely related to the Atlantic cod (Gadus morhua) as is the Pacific cod (Gadus macrocephalus), which suggests that T. chalcogramma and G. macrocephalus represent separate invasions of the Pacific Basin. The Pacific tomcod (Microgadus proximus) is more closely related to the Barents Sea navaga (Eleginus navaga) than to the congeneric Atlantic tomcod (Microgadus tomcod), which suggests that the Pacific species is derived from the Eleginus lineage and that Eleginus should be synonymized with Microgadus. Molecular divergences between each of the three endemic Pacific species and their respective closest relatives are similar and consistent with contemporaneous speciation events following the reopening of the Bering Strait ca. 3.0–3.5 million years BP. In contrast, the Greenland cod (Gadus ogac) and the Pacific cod have essentially identical mtDNA sequences; differences between them are less than those found within G. morhua. The Greenland cod appears to represent a contemporary northward and eastward range extension of the Pacific cod, and should be synonymized with it as G. -

December 20, 2007
BEFORE THE SECRETARY OF COMMERCE PETITION TO LIST THE RIBBON SEAL (HISTRIOPHOCA FASCIATA) AS A THREATENED OR ENDANGERED SPECIES UNDER THE ENDANGERED SPECIES ACT © G. CARLETON RAY CENTER FOR BIOLOGICAL DIVERSITY DECEMBER 20, 2007 Notice of Petition____________________________________________________ Carlos M. Gutierrez Secretary of Commerce U.S. Department of Commerce 1401 Constitution Avenue, N.W., Room 5516 Washington, D.C. 20230 Dr. William Hogarth Assistant Administrator for Fisheries National Oceanographic and Atmospheric Administration 1315 East-West Highway Silver Springs, MD 20910 PETITIONER The Center for Biological Diversity 1095 Market Street, Suite 511 San Francisco, CA 94103 ph: (415) 436-9682 ext 301 fax: (415) 436-9683 __________________________ Date: this 20th day of December, 2007 Shaye Wolf, Ph.D. Martha Palomino Tovar, Ph.D. Candidate Brendan Cummings Center for Biological Diversity Pursuant to Section 4(b) of the Endangered Species Act (“ESA”), 16 U.S.C. §1533(b), Section 553(3) of the Administrative Procedures Act, 5 U.S.C. § 553(e), and 50 C.F.R. §424.14(a), the Center for Biological Diversity (“Petitioner”) hereby petitions the Secretary of Commerce, through the National Marine Fisheries Service (“NMFS”), to list the ribbon seal (Histriophoca fasciata) as a threatened or endangered species and to designate critical habitat to ensure its survival and recovery. The Center for Biological Diversity (“Center”) is a non-profit, public interest environmental organization dedicated to the protection of native species and their habitats through science, policy, and environmental law. The Center has over 40,000 members in Alaska and throughout the United States. The Center and its members are concerned with the conservation of endangered species, including the ribbon seal, and the effective implementation of the ESA. -

Complete Mitochondrial Genome Sequences of the Arctic Ocean Codwshes Arctogadus Glacialis and Boreogadus Saida Reveal Oril and Trna Gene Duplications
Polar Biol (2008) 31:1245–1252 DOI 10.1007/s00300-008-0463-7 ORIGINAL PAPER Complete mitochondrial genome sequences of the Arctic Ocean codWshes Arctogadus glacialis and Boreogadus saida reveal oriL and tRNA gene duplications Ragna Breines · Anita Ursvik · Marianne Nymark · Steinar D. Johansen · Dag H. Coucheron Received: 4 December 2007 / Revised: 16 April 2008 / Accepted: 5 May 2008 / Published online: 27 May 2008 © The Author(s) 2008 Abstract We have determined the complete mitochon- Introduction drial genome sequences of the codWshes Arctogadus gla- cialis and Boreogadus saida (Order Gadiformes, Family More than 375 complete sequenced mitochondrial genomes Gadidae). The 16,644 bp and 16,745 bp mtDNAs, respec- from ray-Wnned Wshes have so far (December 2007) been tively, contain the same set of 37 structural genes found in submitted to the database (http://www.ncbi.nlm.nih.gov), all vertebrates analyzed so far. The gene organization is and many of these sequences have contributed considerably conserved compared to other Gadidae species, but with one to resolving phylogenetic relationships among Wshes. Evo- notable exception. B. saida contains heteroplasmic rear- lutionary relationships at diVerent taxonomic levels have rangement-mediated duplications that include the origin of been addressed, including Division (Inoue et al. 2003; Miya light-strand replication and nearby tRNA genes. Examina- et al. 2003), Subdivision (Ishiguro et al. 2003), Genus tion of the mitochondrial control region of A. glacialis, (Doiron et al. 2002; Minegishi et al. 2005), and Species B. saida, and four additional representative Gadidae genera (Yanagimoto et al. 2004; Ursvik et al. 2007). identiWed a highly variable domain containing tandem The circular mitochondrial genomes from ray-Wnned repeat motifs in A. -

Radioactivity in the Arctic Seas
IAEA-TECDOC-1075 XA9949696 Radioactivity in the Arctic Seas Report for the International Arctic Seas Assessment Project (IASAP) ffl INTERNATIONAL ATOMIC ENERGY AGENCA / Y / 1JrrziZr^AA 30-16 The originating Section of this publication in the IAEA was: Radiometrics Section International Atomic Energy Agency Marine Environment Laboratory B.P. 800 MC 98012 Monaco Cedex RADIOACTIVITY IN THE ARCTIC SEAS IAEA, VIENNA, 1999 IAEA-TECDOC-1075 ISSN 1011-4289 ©IAEA, 1999 Printe IAEe th AustriAn y i d b a April 1999 FOREWORD From 199 o 1993t e Internationa6th l Atomic Energy Agency's Marine Environment Laboratory (IAEA-MEL s engage IAEA'e wa ) th n di s International Arctic Seas Assessment Project (IASAP whicn i ) h emphasi bees ha sn place criticaa n do l revie f environmentawo l conditions in the Arctic Seas. IAEA-MEe Th L programme, organize framewore th n dIASAi e th f ko P included: (i) an oceanographic and an ecological description of the Arctic Seas; provisioe th (ii )centra a f no l database facilitIASAe th r yfo P programm collectione th r efo , synthesi interpretatiod san datf nmarino n ao e radioactivit Arctie th n yi c Seas; (iii) participation in official expeditions to the Kara Sea organized by the joint Russian- Norwegian Experts Group (1992, 1993 and 1994), the Russian Academy of Sciences (1994), and the Naval Research Laboratory and Norwegian Defence Research Establishment (1995); (iv) assistance wit d n laboratorsiti han u y based radiometric measurement f curreno s t radionuclide concentrations in the Kara Sea; (v) organization of analytical quality assurance intercalibration exercises among the participating laboratories; (vi) computer modellin e potentiath f o g l dispersa f radionuclideo l s released froe mth dumped f assessmeno wast d associatee ean th f o t d radiological consequencee th f o s disposals on local, regional and global scales; (vii) in situ and laboratory based assessment of distribution coefficients (Kd) and concentration factor sArctie (CFth r c)fo environment. -

Humboldt Bay Fishes
Humboldt Bay Fishes ><((((º>`·._ .·´¯`·. _ .·´¯`·. ><((((º> ·´¯`·._.·´¯`·.. ><((((º>`·._ .·´¯`·. _ .·´¯`·. ><((((º> Acknowledgements The Humboldt Bay Harbor District would like to offer our sincere thanks and appreciation to the authors and photographers who have allowed us to use their work in this report. Photography and Illustrations We would like to thank the photographers and illustrators who have so graciously donated the use of their images for this publication. Andrey Dolgor Dan Gotshall Polar Research Institute of Marine Sea Challengers, Inc. Fisheries And Oceanography [email protected] [email protected] Michael Lanboeuf Milton Love [email protected] Marine Science Institute [email protected] Stephen Metherell Jacques Moreau [email protected] [email protected] Bernd Ueberschaer Clinton Bauder [email protected] [email protected] Fish descriptions contained in this report are from: Froese, R. and Pauly, D. Editors. 2003 FishBase. Worldwide Web electronic publication. http://www.fishbase.org/ 13 August 2003 Photographer Fish Photographer Bauder, Clinton wolf-eel Gotshall, Daniel W scalyhead sculpin Bauder, Clinton blackeye goby Gotshall, Daniel W speckled sanddab Bauder, Clinton spotted cusk-eel Gotshall, Daniel W. bocaccio Bauder, Clinton tube-snout Gotshall, Daniel W. brown rockfish Gotshall, Daniel W. yellowtail rockfish Flescher, Don american shad Gotshall, Daniel W. dover sole Flescher, Don stripped bass Gotshall, Daniel W. pacific sanddab Gotshall, Daniel W. kelp greenling Garcia-Franco, Mauricio louvar -

Cardiophysiological Responses of the Air-Breathing Alaska Blackfish to Cold Acclimation and Chronic Hypoxic Submergence at 5°C Jonathan A
© 2020. Published by The Company of Biologists Ltd | Journal of Experimental Biology (2020) 223, jeb225730. doi:10.1242/jeb.225730 RESEARCH ARTICLE Cardiophysiological responses of the air-breathing Alaska blackfish to cold acclimation and chronic hypoxic submergence at 5°C Jonathan A. W. Stecyk1,‡, Christine S. Couturier1,*, Denis V. Abramochkin2,3,4,*, Diarmid Hall1, Asia Arrant-Howell1, Kerry L. Kubly1, Shyanne Lockmann1, Kyle Logue1, Lenett Trueblood1, Connor Swalling1, Jessica Pinard1 and Angela Vogt1 ABSTRACT 2016). Consequently, in winter, cardiac physiology must be The Alaska blackfish (Dallia pectoralis) remains active at cold adjusted to accommodate the cold temperature-driven effects on temperatures when experiencing aquatic hypoxia without air access. contractility, blood viscosity and associated vascular resistance to To discern the cardiophysiological adjustments that permit this ensure the efficient transport of respiratory gases, nutrients, waste behaviour, we quantified the effect of acclimation from 15°C to 5°C in products and signalling molecules of the endocrine system among normoxia (15N and 5N fish), as well as chronic hypoxic submergence tissues via the circulatory system (Young and Egginton, 2011). In (6–8 weeks; ∼6.3–8.4 kPa; no air access) at 5°C (5H fish), on in vivo addition, in winter, cardiac electrical excitability must be adjusted to coincide with temperature-dependent reductions in heart rate ( fH), and spontaneous heart rate ( fH), electrocardiogram, ventricular action potential (AP) shape and duration (APD), the background inward while maintaining cardiac excitability and protecting against cardiac + arrhythmias that can be induced by cold temperature and oxygen rectifier (IK1) and rapid delayed rectifier (IKr)K currents and ventricular gene expression of proteins involved in excitation–contraction deprivation (Vornanen, 2017). -

Biodiversity of Arctic Marine Fishes: Taxonomy and Zoogeography
Mar Biodiv DOI 10.1007/s12526-010-0070-z ARCTIC OCEAN DIVERSITY SYNTHESIS Biodiversity of arctic marine fishes: taxonomy and zoogeography Catherine W. Mecklenburg & Peter Rask Møller & Dirk Steinke Received: 3 June 2010 /Revised: 23 September 2010 /Accepted: 1 November 2010 # Senckenberg, Gesellschaft für Naturforschung and Springer 2010 Abstract Taxonomic and distributional information on each Six families in Cottoidei with 72 species and five in fish species found in arctic marine waters is reviewed, and a Zoarcoidei with 55 species account for more than half list of families and species with commentary on distributional (52.5%) the species. This study produced CO1 sequences for records is presented. The list incorporates results from 106 of the 242 species. Sequence variability in the barcode examination of museum collections of arctic marine fishes region permits discrimination of all species. The average dating back to the 1830s. It also incorporates results from sequence variation within species was 0.3% (range 0–3.5%), DNA barcoding, used to complement morphological charac- while the average genetic distance between congeners was ters in evaluating problematic taxa and to assist in identifica- 4.7% (range 3.7–13.3%). The CO1 sequences support tion of specimens collected in recent expeditions. Barcoding taxonomic separation of some species, such as Osmerus results are depicted in a neighbor-joining tree of 880 CO1 dentex and O. mordax and Liparis bathyarcticus and L. (cytochrome c oxidase 1 gene) sequences distributed among gibbus; and synonymy of others, like Myoxocephalus 165 species from the arctic region and adjacent waters, and verrucosus in M. scorpius and Gymnelus knipowitschi in discussed in the family reviews. -

Creating Sense from Non-Sense Dna: De Novo Genesis and Evolutionary History of Antifreeze Glycoprotein Gene in Northern Cod Fishes (Gadidae)
CREATING SENSE FROM NON-SENSE DNA: DE NOVO GENESIS AND EVOLUTIONARY HISTORY OF ANTIFREEZE GLYCOPROTEIN GENE IN NORTHERN COD FISHES (GADIDAE) BY XUAN ZHUANG DISSERTATION Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biology with a concentration in Ecology, Ethology, and Evolution in the Graduate College of the University of Illinois at Urbana-Champaign, 2013 Urbana, Illinois Doctoral Committee: Professor Chi-Hing Christina Cheng, Chair, Director of Research Professor Arthur L. DeVries Professor Gary J. Olsen Associate Professor Kurt E. Kwast ABSTRACT Gadids (cod fishes) are ecologically prominent and important species of the north polar marine fauna. Seven gadid species are known to have evolved antifreeze glycoproteins (AFGPs) that enable their survival from inoculative freezing by environmental ice in frigid Arctic and sub- Arctic waters. AFGPs protect the fish from freezing death by binding to ice crystals and arresting ice growth within the fish. How and from where this important new genetic trait evolved in the gadid genome remained unknown, as thus far there are no homologous sequences in databases to infer genetic ancestry based on sequence homology. Additionally, the seven AFGP-bearing species are dispersed in separate clades of the gadid phylogeny, indicating a complicated evolutionary history of AFGP in the gadid lineage. The AFGP loci in three gadid species were isolated and characterized to investigate the genetic origin and evolutionary pathway of the AFGP gene family in the gadid lineage. Two of these species are polar cod Boreogadus saida and Atlantic tomcod Microgadus tomcod, which occur in distinct clades representing two possible separate origins of the gadid AFGP. -
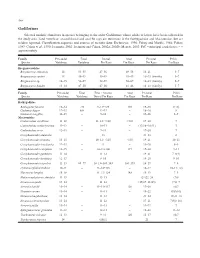
Gadiformes Selected Meristic Characters in Species Belonging to the Order Gadiformes Whose Adults Or Larvae Have Been Collected in the Study Area
548 Gadiformes Selected meristic characters in species belonging to the order Gadiformes whose adults or larvae have been collected in the study area. Total vertebrae, second dorsal and anal fin rays are numerous in the Bathygadidae and Macrouridae, but are seldom reported. Classification sequence and sources of meristic data: Eschmeyer, 1990; Fahay and Markle, 1984; Fahay, 1989; Cohen et al., 1990; Iwamoto, 2002; Iwamoto and Cohen, 2002a; 2002b; Merrett, 2003. PrC = principal caudal rays; ~ = approximately Family Precaudal Total Dorsal Anal Pectoral Pelvic Species Vertebrae Vertebrae Fin Rays Fin Rays Fin Rays Fin Rays Bregmacerotidae Bregmaceros atlanticus 14 53–55 47–56 49–58 16–21 5–7 Bregmaceros cantori 14 45–49 45–49 45–49 16–23 (family) 5–7 Bregmaceros sp. 14–15 52–59 52–59 58–69 16–23 (family) 5–7 Bregmaceros houdei 13–14 47–50 47–50 41–46 16–23 (family) 5–7 Family Precaudal Total First + Second Anal Pectoral Pelvic Species Vertebrae Vertebrae Dorsal Fin Rays Fin Rays Fin Rays Fin Rays Bathygadidae Bathygadus favosus 12–14 ~70 9–11+125 110 15–18 9(10) Gadomus dispar 12–13 80+ 12–13 – 18–20 8 Gadomus longifilis 11–13 – 9–11 – 14–16 8–9 Macrouridae Caelorinchus caribbeus 11–12 – 11–12+>110 >110 17–20 7 Caelorinchus coelorhynchus 11–12 – 10–11 – (17)18–20(21) 7 Caelorinchus occa 12–13 – 9–11 – 17–20 7 Coryphaenoides alateralis – 13 – 21–23 8 Coryphaenoides armatus 13–15 – 10–12+~125 ~135 19–21 10–11 Coryphaenoides brevibarbis 12–13 – 9 – 19–20 8–9 Coryphaenoides carapinus 12–15 – 10–11+100 117 17–20 9–11 Coryphaenoides guentheri -

Descriptive Key to the Otoliths of Gadid Fishes of the Bering, Chukchi, and Beaufort Seas KATHRYN J
ARCTIC VOL. 34, NO. 1 (MARCH 1981), P. 55-59 Descriptive Key to the Otoliths of Gadid Fishes of the Bering, Chukchi, and Beaufort Seas KATHRYN J. FROST’ ABSTRACT. An illustrated key with supplementary descriptive material is presented for six species or species groups of gadid fishes which are of trophic importance in the Bering, Chukchi, and Beaufort seas. These species include: Arcrogadus spp. Djagin, Boreogadus saida (Lepechin), Eleginus gracilis (Tilesius), Gadus macrocephalus Tilesius, Microgadus proximus (Girard), and Theragra chalcogramma (Pallas). RESUME. Une clC d’identificationillustree par des figures avec un complement descriptif est ici presentee pour six espbces ou groupes d’espbces de poissons de la famiile des gadidCs, lesquels ont une importance au point de vue trophique dans les mers de BCring, des Tchouktches et deBeaufort. Ces espbcescomprennent: Arctogadus spp. Djagin, Boreogadus saida (Lepechin), Eleginus gracilis (Tilesius), Gadus macrocephalus Tilesius, Microgadus proximus (Girard), et Theragra chalcogramma (Pallas). Traduit par Jean-Guy Brossard, Laboratoire d’Archeologie de I’Universit6 due Quebec A Montreal. Key words: otolith, gadid fishes, Arctogadus, Boreogadus, Eleginus, Gadus, Microgadus, Theragra INTRODUCTION size or when certain features vary such that an otolith of Investigations of food habits of marine animals almost one species closelyresembles that of other species. Furth- invariably involve analysisof stomach contents. Success- er, keys,are often usedby readers who have little familiar- ful stomach contents analysis usually requires that prey ity with otoliths and limited access to comparative mate- items be recognized by characteristic fragments. In this rial, and who therefore require more detailed descriptive respect the sagittal otoliths of bony fishes are very useful material. -

Fishes-Of-The-Salish-Sea-Pp18.Pdf
NOAA Professional Paper NMFS 18 Fishes of the Salish Sea: a compilation and distributional analysis Theodore W. Pietsch James W. Orr September 2015 U.S. Department of Commerce NOAA Professional Penny Pritzker Secretary of Commerce Papers NMFS National Oceanic and Atmospheric Administration Kathryn D. Sullivan Scientifi c Editor Administrator Richard Langton National Marine Fisheries Service National Marine Northeast Fisheries Science Center Fisheries Service Maine Field Station Eileen Sobeck 17 Godfrey Drive, Suite 1 Assistant Administrator Orono, Maine 04473 for Fisheries Associate Editor Kathryn Dennis National Marine Fisheries Service Offi ce of Science and Technology Fisheries Research and Monitoring Division 1845 Wasp Blvd., Bldg. 178 Honolulu, Hawaii 96818 Managing Editor Shelley Arenas National Marine Fisheries Service Scientifi c Publications Offi ce 7600 Sand Point Way NE Seattle, Washington 98115 Editorial Committee Ann C. Matarese National Marine Fisheries Service James W. Orr National Marine Fisheries Service - The NOAA Professional Paper NMFS (ISSN 1931-4590) series is published by the Scientifi c Publications Offi ce, National Marine Fisheries Service, The NOAA Professional Paper NMFS series carries peer-reviewed, lengthy original NOAA, 7600 Sand Point Way NE, research reports, taxonomic keys, species synopses, fl ora and fauna studies, and data- Seattle, WA 98115. intensive reports on investigations in fi shery science, engineering, and economics. The Secretary of Commerce has Copies of the NOAA Professional Paper NMFS series are available free in limited determined that the publication of numbers to government agencies, both federal and state. They are also available in this series is necessary in the transac- exchange for other scientifi c and technical publications in the marine sciences. -

The Ringed Seal, Phoca Hispida, of the Canadian Western Arctic Jimmy Memorana of Holman, Northwest Territories, with a Day's Catch of Ringed Seals
Canadian Bulletin of Fisheries_and Aquatic-Sciences 216 DFO - Lib ary / MPO - B b otheque 11111 I 1111 11111 11 11 12039511 The Ringed Seal, Phoca hispida. of the Canadian Western Arctic Thomas G. Smith -SW D23 92/3 2 C. / Fisheries Pêches 1+ and Oceans et Océans The Ringed Seal, Phoca hispida, of the Canadian Western Arctic Jimmy Memorana of Holman, Northwest Territories, with a day's catch of ringed seals. Il Canadian Bulletin of Fisheries and Aquatic Sciences 216 The Ringed Seal, Phoca hispida, of the Canadian Western Arctic Thomas G. Smith Department of Fisheries and Oceans Arctic Biological Station 555 St-Pierre Blvd. Sainte-Anne-de-Bellevue, Quebec H9X 3R4 DEPARTMENT OF FISHERIES AND OCEANS Ottawa 1987 III The Canadian Bulletins of Fisheries and Aquatic. Sciences are designed to interpret current knowledge in scientific fields pertinent to Canadian fisheries and aquatic environments. The Canadian Journal of Fisheries and Aquatic Sciences is published in annual volumes of monthly issues. Canadian Special Publications of Fisheries and Aquatic Sciences arc issued periodically. These series are available from authorized bookstore agents and other bookstores, or you may send your prepaid order to the Canadian Government Publishing Centre, Supply and Services Canada, Ottawa, Ont. KIA 0S9. Make cheque or money order payable in Canadian funds to the Receiver General for Canada. Information and Publications Branch Dixi Lambert Director General Johanna M. Reinhart, M.Sc. Director and Editor Gerald J. Neville Editorial and Publishing Services Editorial Office: Department of Fisheries and Oceans Communications Directorate Information and Publications Branch 200 Kent Street Ottawa, Ontario, Canada K IA 0E6 Typesetter: K.G.