Co-Chaperones in Targeting and Delivery of Misfolded Proteins to the 26S Proteasome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Control of Protein Function
3 Control of Protein Function In the cell, precise regulation of protein function is essential to avoid chaos. This chapter describes the most important molecular mechanisms by which protein function is regulated in cells. These range from control of a protein’s location and lifetime within the cell to the binding of regulatory molecules and covalent modifications such as phosphorylation that rapidly switch protein activity on or off. Also covered here are the nucleotide-driven switches in conformation that underlie the action of motor proteins and that regulate many signal transduction pathways. 3-0 Overview: Mechanisms of Regulation 3-1 Protein Interaction Domains 3-2 Regulation by Location 3-3 Control by pH and Redox Environment 3-4 Effector Ligands: Competitive Binding and Cooperativity 3-5 Effector Ligands: Conformational Change and Allostery 3-6 Protein Switches Based on Nucleotide Hydrolysis 3-7 GTPase Switches: Small Signaling G Proteins 3-8 GTPase Switches: Signal Relay by Heterotrimeric GTPases 3-9 GTPase Switches: Protein Synthesis 3-10 Motor Protein Switches 3-11 Regulation by Degradation 3-12 Control of Protein Function by Phosphorylation 3-13 Regulation of Signaling Protein Kinases: Activation Mechanism 3-14 Regulation of Signaling Protein Kinases: Cdk Activation 3-15 Two-Component Signaling Systems in Bacteria 3-16 Control by Proteolysis: Activation of Precursors 3-17 Protein Splicing: Autoproteolysis by Inteins 3-18 Glycosylation 3-19 Protein Targeting by Lipid Modifications 3-20 Methylation, N-acetylation, Sumoylation and Nitrosylation 3-0 Overview: Mechanisms of Regulation Protein function in living cells is precisely regulated A typical bacterial cell contains a total of about 250,000 protein molecules (comprising different amounts of each of several thousand different gene products), which are packed into a volume so small that it has been estimated that, on average, they are separated from one another by a distance that would contain only a few molecules of water. -

UBQLN2 Polyclonal Antibody C-Terminal Ubiquitin-Associated Domain
UBQLN2 polyclonal antibody C-terminal ubiquitin-associated domain. They physically associate with both proteasomes and ubiquitin ligases, Catalog Number: PAB1699 and thus thought to functionally link the ubiquitination machinery to the proteasome to affect in vivo protein Regulatory Status: For research use only (RUO) degradation. This ubiquilin has also been shown to bind the ATPase domain of the Hsp70-like Stch protein. Product Description: Rabbit polyclonal antibody raised [provided by RefSeq] against synthetic peptide of UBQLN2. References: Immunogen: A synthetic peptide (conjugated with KLH) 1. Structural studies of the interaction between ubiquitin corresponding to C-terminus of human UBQLN2. family proteins and proteasome subunit S5a. Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Host: Rabbit Biochemistry. 2002 Feb 12;41(6):1767-77. 2. The hPLIC proteins may provide a link between the Reactivity: Human ubiquitination machinery and the proteasome. Kleijnen Applications: WB-Ce MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha (See our web site product page for detailed applications NL, Gill G, Howley PM. Mol Cell. 2000 Aug;6(2):409-19. information) 3. Selection system for genes encoding nuclear-targeted proteins. Ueki N, Oda T, Kondo M, Yano K, Noguchi T, Protocols: See our web site at Muramatsu M. Nat Biotechnol. 1998 http://www.abnova.com/support/protocols.asp or product Dec;16(13):1338-42. page for detailed protocols Form: Liquid Purification: Protein G purification Recommend Usage: Western Blot (1:1000) The optimal working dilution should be determined by the end user. Storage Buffer: In PBS (0.09% sodium azide) Storage Instruction: Store at 4°C. -

1519038862M28translationand
Paper No. : 15 Molecular Cell Biology Module : 28 Translation and Post-translation Modifications in Eukaryotes Development Team Principal Investigator : Prof. Neeta Sehgal Department of Zoology, University of Delhi Co-Principal Investigator : Prof. D.K. Singh Department of Zoology, University of Delhi Paper Coordinator : Prof. Kuldeep K. Sharma Department of Zoology, University of Jammu Content Writer : Dr. Renu Solanki, Deen Dayal Upadhyaya College Dr. Sudhida Gautam, Hansraj College, University of Delhi Mr. Kiran K. Salam, Hindu College, University of Delhi Content Reviewer : Prof. Rup Lal Department of Zoology, University of Delhi 1 Molecular Genetics ZOOLOGY Translation and Post-translation Modifications in Eukaryotes Description of Module Subject Name ZOOLOGY Paper Name Molecular Cell Biology; Zool 015 Module Name/Title Cell regulatory mechanisms Module Id M28: Translation and Post-translation Modifications in Eukaryotes Keywords Genome, Proteome diversity, post-translational modifications, glycosylation, phosphorylation, methylation Contents 1. Learning Objectives 2. Introduction 3. Purpose of post translational modifications 4. Post translational modifications 4.1. Phosphorylation, the addition of a phosphate group 4.2. Methylation, the addition of a methyl group 4.3. Glycosylation, the addition of sugar groups 4.4. Disulfide bonds, the formation of covalent bonds between 2 cysteine amino acids 4.5. Proteolysis/ Proteolytic Cleavage 4.6. Subunit binding to form a multisubunit protein 4.7. S-nitrosylation 4.8. Lipidation 4.9. Acetylation 4.10. Ubiquitylation 4.11. SUMOlytion 4.12. Vitamin C-Dependent Modifications 4.13. Vitamin K-Dependent Modifications 4.14. Selenoproteins 4.15. Myristoylation 5. Chaperones: Role in PTM and mechanism 6. Role of PTMs in diseases 7. Detecting and Quantifying Post-Translational Modifications 8. -

Protein Targeting Or Protein Sorting
Protein targeting or Protein sorting Refer Page 1068 to 1074 Principles of Biochemistry by Lehninger & Page 663 Baltimore Mol Cell Biology • Protein targeting or Protein sorting is the process of delivery of newly synthesized proteins to their proper cellular destinations • Proteins are sorted to the endoplasmic reticulum (ER), mitochondria, chloroplasts, lysosomes, peroxisomes, and the nucleus by different mechanisms • The process can occur either during protein synthesis or soon after synthesis of proteins by translation at the ribosome. • Most of the integral membrane proteins, secretory proteins and lysosomal proteins are sorted to ER lumen from where these proteins are modified for further sorting • For membrane proteins, targeting leads to insertion of the protein into the lipid bilayer • For secretory/water-soluble proteins, targeting leads to translocation of the entire protein across the membrane into the aqueous interior of the organelle. • Protein destined for cytosol simply remain where they are synthesized • Mitochondrial and chloroplast proteins are first completely synthesized and released from ribosomes. These are then bound by cytosolic chaperone proteins and delivered to receptor on target organelle. • Nuclear proteins such as DNA and RNA polymerases, histones, topoisomerases and proteins that regulate gene expression contain Nuclear localization signal (NLS) which is not removed after the protein is translocated. Unlike ER localization signal sequence which is at N terminal, NLS ca be located almost anywhere along the -

Targeting the Ubiquitin System in Glioblastoma', Frontiers in Oncology
Citation for published version: Licchesi, J 2020, 'Targeting the Ubiquitin System in Glioblastoma', Frontiers in Oncology. https://doi.org/10.3389/fonc.2020.574011 DOI: 10.3389/fonc.2020.574011 Publication date: 2020 Document Version Publisher's PDF, also known as Version of record Link to publication University of Bath Alternative formats If you require this document in an alternative format, please contact: [email protected] General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 24. Sep. 2021 REVIEW published: 25 November 2020 doi: 10.3389/fonc.2020.574011 Targeting the Ubiquitin System in Glioblastoma Nico Scholz 1, Kathreena M. Kurian 2, Florian A. Siebzehnrubl 3 and Julien D. F. Licchesi 1* 1 Department of Biology & Biochemistry, University of Bath, Bath, United Kingdom, 2 Brain Tumour Research Group, Institute of Clinical Neurosciences, University of Bristol, Bristol, United Kingdom, 3 Cardiff University School of Biosciences, European Cancer Stem Cell Research Institute, Cardiff, United Kingdom Glioblastoma is the most common primary brain tumor in adults with poor overall outcome and 5-year survival of less than 5%. Treatment has not changed much in the last decade or so, with surgical resection and radio/chemotherapy being the main options. -

(Stch) Gene Reduces Prion Disease Incubation Time in Mice
Overexpression of the Hspa13 (Stch) gene reduces prion disease incubation time in mice Julia Grizenkovaa,b, Shaheen Akhtara,b, Holger Hummericha,b, Andrew Tomlinsona,b, Emmanuel A. Asantea,b, Adam Wenborna,b, Jérémie Fizeta,b, Mark Poultera,b, Frances K. Wisemanb, Elizabeth M. C. Fisherb, Victor L. J. Tybulewiczc, Sebastian Brandnerb, John Collingea,b, and Sarah E. Lloyda,b,1 aMedical Research Council (MRC) Prion Unit and bDepartment of Neurodegenerative Disease, University College London (UCL) Institute of Neurology, London WC1N 3BG, United Kingdom; and cDivision of Immune Cell Biology, MRC National Institute for Medical Research, London NW7 1AA, United Kingdom Edited by Reed B. Wickner, National Institutes of Health, Bethesda, MD, and approved July 10, 2012 (received for review May 28, 2012) Prion diseases are fatal neurodegenerative disorders that include way crosses and a heterogeneous cross (12–14). In these studies, bovine spongiform encephalopathy (BSE) and scrapie in animals the underlying functional polymorphism is unknown but could be and Creutzfeldt-Jakob disease (CJD) in humans. They are character- an amino acid change within the coding region of a protein or ized by long incubation periods, variation in which is determined by splicing variant or may occur within noncoding sequences such as many factors including genetic background. In some cases it is pos- untranslated regions, promoters, or other regulatory regions sible that incubation time may be directly correlated to the level of thereby influencing the pattern or level of expression. Although gene expression. To test this hypothesis, we combined incubation incubation time is a polygenic trait, it is possible that in some cases, time data from five different inbred lines of mice with quantitative there may be a direct correlation between incubation time and gene expression profiling in normal brains and identified five genes individual gene expression level. -

Development and Validation of a Serum Biomarker Panel for The
Journal of Cancer 2017, Vol. 8 2346 Ivyspring International Publisher Journal of Cancer 2017; 8(12): 2346-2355. doi: 10.7150/jca.19465 Research Paper Development and Validation of a Serum Biomarker Panel for the Detection of Esophageal Squamous Cell Carcinoma through RNA Transcriptome Sequencing Shan Xing1,2†, Xin Zheng1,2†, Li-Qiang Wei4†, Shi-Jian Song5, Dan Liu1,3, Ning Xue1,3, Xiao-Min Liu1,2, Mian-Tao Wu1,2, Qian Zhong1,3, Chu-Mei Huang6, Mu-Sheng Zeng1,3, Wan-Li Liu1,2 1. State Key Laboratory of Oncology in Southern China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China 2. Department of Clinical Laboratory, Sun Yat-sen University Cancer Center, Guangzhou, China 3. Department of Experimental Research, Sun Yat-sen University cancer center, Guangzhou, China 4. Department of Clinical Laboratory, Shaanxi Provincial People’s Hospital, Xian, China 5. Guangdong Experimental High School, Guangzhou, China 6. Department of Laboratory Medicine, Sun Yat-sen University First Affiliated Hospital, Guangzhou, China † Xing S, Zheng X and Wei LQ contributed equally to this work Corresponding authors: Wan-Li Liu. Email: [email protected]; Phone: 86-020-87343196. Department of Clinical Laboratory, Sun Yat-sen University Cancer Center, 651 Dongfeng Road East,Guangzhou 510060, P.R. China. Mu-Sheng Zeng; Email: [email protected] Phone: +86-020-87343192. Department of Experimental Research, Sun Yat-sen University cancer center, 651 Dongfeng Road East,Guangzhou 510060, P.R.China © Ivyspring International Publisher. This is an open access article distributed under the terms of the Creative Commons Attribution (CC BY-NC) license (https://creativecommons.org/licenses/by-nc/4.0/). -

Hsp70 in Cancer: Partner Or Traitor to Immune System
REVIEW ARTICLE Iran J Allergy Asthma Immunol December 2019; 18(6):589-604. Hsp70 in Cancer: Partner or Traitor to Immune System Mehdi Asghari Vostakolaei1,2, Jalal Abdolalizadeh1,3, Mohammad Saeid Hejazi2,4,5, Shirafkan Kordi6, Ommoleila Molavi2,7 1 Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran 2 Department of Pharmaceutical Biotechnology, Faculty of pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran 3 Paramedical Faculty, Tabriz University of Medical Sciences, Tabriz, Iran 4 Department of Molecular Medicine, School of Advanced Biomedical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran 5 Molecular Medicine Research Center, Biomedicine Institute, Tabriz University of Medical Sciences, Tabriz, Iran 6 Department of Medical Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran 7 Biotechnology Research Center, Faculty of Pharmacy, Tabriz University of Medical Sciences, Tabriz, Iran Received: 19 August 2019; Received in revised form: 5 September 2019; Accepted: 14 September 2019 ABSTRACT Heat shock protein 70.1 (Hsp70.1), also known as Hsp70, is a highly conserved member of the heat shock protein family that exists in all living organisms and determines the protein fate as molecular chaperones. Hsp70 basal expression is undetectable or low in most unstressed normal cells, however, its abundant presence in several types of human cancer cells is reported. Several studies support upregulated Hsp70 involved in tumor progression and drug resistance through modulation of cell death pathways and suppresses anticancer immune responses. However, numerous studies have confirmed that Hsp70 can also induce anticancer immune responses through the activation of immune cells in particular antigen-presenting cells (APCs). -

RTL8 Promotes Nuclear Localization of UBQLN2 to Subnuclear Compartments Associated with Protein Quality Control
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.21.440788; this version posted April 22, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Title: RTL8 promotes nuclear localization of UBQLN2 to subnuclear compartments associated with protein quality control Harihar Milaganur Mohan1,2, Amit Pithadia1, Hanna Trzeciakiewicz1, Emily V. Crowley1, Regina Pacitto1 Nathaniel Safren1,3, Chengxin Zhang4, Xiaogen Zhou4, Yang Zhang4, Venkatesha Basrur5, Henry L. Paulson1,*, Lisa M. Sharkey1,* 1. Department of Neurology, University of Michigan, Ann Arbor, MI 48109-2200 2. Graduate Program in Cellular and Molecular Biology, University of Michigan, Ann Arbor, MI 48109- 2200 3. Present address: Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL 60611 4. Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109-2200 5. Department of Pathology, University of Michigan Medical School, Ann Arbor, MI 48109. *Corresponding authors: Henry Paulson: Department of Neurology, University of Michigan, Ann Arbor, MI 48109-2200; [email protected]; Tel. (734) 615-5632; Fax. (734) 615-5655 Lisa M Sharkey: Department of Neurology, University of Michigan, Ann Arbor, MI 48109-2200; [email protected]; Tel. (734) 763-3496; Fax (734) 615-5655 Keywords: Ubiquilin, UBQLN2, RTL8, Nuclear Protein Quality Control, Ubiquitin Proteasome System Declarations Funding: This work was supported by NIH 9R01NS096785-06, 1P30AG053760-01, The Amyotrophic Lateral Sclerosis Foundation and the UM Protein Folding Disease Initiative. Conflicts of interest/Competing interests: None declared Availability of data and material: • The authors confirm that the data supporting the findings in this are available within the article, at repository links provided within the article, and within its supplementary files. -

Differential Effects of STCH and Stress-Inducible Hsp70 on the Stability and Maturation of NKCC2
International Journal of Molecular Sciences Article Differential Effects of STCH and Stress-Inducible Hsp70 on the Stability and Maturation of NKCC2 Dalal Bakhos-Douaihy 1,2, Elie Seaayfan 1,2,† , Sylvie Demaretz 1,2,†, Martin Komhoff 3,‡ and Kamel Laghmani 1,2,*,‡ 1 Centre de Recherche des Cordeliers, INSERM, Sorbonne Université, USPC, Université Paris Descartes, Université Paris Diderot, 75006 Paris, France; [email protected] (D.B.-D.); [email protected] (E.S.); [email protected] (S.D.) 2 CNRS, ERL8228, 75006 Paris, France 3 University Children’s Hospital, Philipps-University, 35043 Marburg, Germany; [email protected] * Correspondence: [email protected]; Tel.: +33-1-44-27-50-68; Fax: +33-1-44-27-51-19 † These authors contributed equally to this work. ‡ These authors contributed equally to this work. Abstract: Mutations in the Na-K-2Cl co-transporter NKCC2 lead to type I Bartter syndrome, a life- threatening kidney disease. We previously showed that export from the ER constitutes the limiting step in NKCC2 maturation and cell surface expression. Yet, the molecular mechanisms involved in this process remain obscure. Here, we report the identification of chaperone stress 70 protein (STCH) and the stress-inducible heat shock protein 70 (Hsp70), as two novel binding partners of the ER-resident form of NKCC2. STCH knock-down increased total NKCC2 expression whereas Hsp70 knock-down or its inhibition by YM-01 had the opposite effect. Accordingly, overexpressing of STCH and Hsp70 exerted opposite actions on total protein abundance of NKCC2 and its folding mutants. -
Download Validation Data
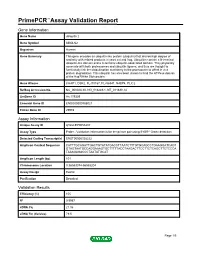
PrimePCR™Assay Validation Report Gene Information Gene Name ubiquilin 2 Gene Symbol UBQLN2 Organism Human Gene Summary This gene encodes an ubiquitin-like protein (ubiquilin) that shares high degree of similarity with related products in yeast rat and frog. Ubiquilins contain a N-terminal ubiquitin-like domain and a C-terminal ubiquitin-associated domain. They physically associate with both proteasomes and ubiquitin ligases; and thus are thought to functionally link the ubiquitination machinery to the proteasome to affect in vivo protein degradation. This ubiquilin has also been shown to bind the ATPase domain of the Hsp70-like Stch protein. Gene Aliases CHAP1, DSK2, FLJ10167, FLJ56541, N4BP4, PLIC2 RefSeq Accession No. NC_000023.10, NG_016249.1, NT_011630.14 UniGene ID Hs.179309 Ensembl Gene ID ENSG00000188021 Entrez Gene ID 29978 Assay Information Unique Assay ID qHsaCEP0055207 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000338222 Amplicon Context Sequence CATTTGCAGATTGACTGTATATGACCTTAATCTTTGTGCAGCCTGAAGGATCAGT GTAGTAATGCCAGGAAAGTGCTTTTTACCTAAGACTTCCTTCTCAGCTTCTCCCA TAAAGAGACCCTAATATGCAT Amplicon Length (bp) 101 Chromosome Location X:56593074-56593204 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 105 R2 0.9967 cDNA Cq 21.06 cDNA Tm (Celsius) 79.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 25.2 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay -

The Heat-Shock Protein/Chaperone Network and Multiple Stress Resistance Pierre Jacob, Heribert Hirt, Abdelhafid Bendahmane
The heat-shock protein/chaperone network and multiple stress resistance Pierre Jacob, Heribert Hirt, Abdelhafid Bendahmane To cite this version: Pierre Jacob, Heribert Hirt, Abdelhafid Bendahmane. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnology Journal, Wiley, 2017, 15 (4), pp.405-414. 10.1111/pbi.12659. hal-01602732 HAL Id: hal-01602732 https://hal.archives-ouvertes.fr/hal-01602732 Submitted on 27 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Plant Biotechnology Journal (2017) 15, pp. 405–414 doi: 10.1111/pbi.12659 Review article The heat-shock protein/chaperone network and multiple stress resistance Pierre Jacob1, Heribert Hirt2,* and Abdelhafid Bendahmane1,* 1Institute of Plant Science—Paris-Saclay, Orsay, France 2Center for Desert Agriculture, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia Received 29 July 2016; Summary revised 25 October 2016; Crop yield has been greatly enhanced during the last century. However, most elite cultivars are accepted 3 November 2016. adapted to temperate climates and are not well suited to more stressful conditions. In the *Correspondence (Tel +331 69 15 33 30; context of climate change, stress resistance is a major concern.