Modulators of Symbiotic Outcome in Sinorhizobium Meliloti
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Elucidation of the Specificity of Sinorhizobium Meliloti Chemoreceptors
Elucidation of the Specificity of Sinorhizobium meliloti Chemoreceptors for Host Derived Attractants Benjamin Allen Webb Dissertation submitted to the faculty of the Virginia Polytechnic Institute and State University in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biological Sciences Birgit E. Scharf, Chair Richard F. Helm Florian D. Schubot Dorothea Tholl Mark A. Williams August 12th, 2016 Blacksburg, VA Keywords: alfalfa, chemotaxis, ligand, methyl accepting chemotaxis protein, symbiosis Copyright © 2016, Benjamin Allen Webb [Type here] Elucidation of the Specificity of S. meliloti Chemoreceptors for Host Derived Attractants Benjamin A. Webb ABSTRACT The bacterium Sinorhizobium (Ensifer) meliloti is a member of the Rhizobiaceae family and can enter a mutualistic, diazotrophic relationship with most plants of the genera Medicago, Melilotus, and Trigonella. Medicago sativa (alfalfa) is an agriculturally important legume that hosts S. meliloti and allows the bacterium to invade the plant root and begin fixing nitrogen. Prior to invasion, S. meliloti exists as a free living bacterium and must navigate through the soil to find alfalfa, using chemical signals secreted by the root. Alfalfa is the 4th most cultivated crop in the United States, therefore, identification of plant host signals that lure S. meliloti, and identification of the bacterium’s chemoreceptors that perceive the signals can aid in propagating the symbiosis more efficiently, thus leading to greater crop yields. Investigations here focus on discovering alfalfa derived attractant signals and matching them to their respective chemoreceptors in S. meliloti. We have determined the chemotactic potency of alfalfa seed exudate and characterized and quantified two classes of attractant compounds exuded by germinating alfalfa seeds, namely, amino acids and quaternary ammonium compounds (QACs). -

Atlas of the Flora of New England: Fabaceae
Angelo, R. and D.E. Boufford. 2013. Atlas of the flora of New England: Fabaceae. Phytoneuron 2013-2: 1–15 + map pages 1– 21. Published 9 January 2013. ISSN 2153 733X ATLAS OF THE FLORA OF NEW ENGLAND: FABACEAE RAY ANGELO1 and DAVID E. BOUFFORD2 Harvard University Herbaria 22 Divinity Avenue Cambridge, Massachusetts 02138-2020 [email protected] [email protected] ABSTRACT Dot maps are provided to depict the distribution at the county level of the taxa of Magnoliophyta: Fabaceae growing outside of cultivation in the six New England states of the northeastern United States. The maps treat 172 taxa (species, subspecies, varieties, and hybrids, but not forms) based primarily on specimens in the major herbaria of Maine, New Hampshire, Vermont, Massachusetts, Rhode Island, and Connecticut, with most data derived from the holdings of the New England Botanical Club Herbarium (NEBC). Brief synonymy (to account for names used in standard manuals and floras for the area and on herbarium specimens), habitat, chromosome information, and common names are also provided. KEY WORDS: flora, New England, atlas, distribution, Fabaceae This article is the eleventh in a series (Angelo & Boufford 1996, 1998, 2000, 2007, 2010, 2011a, 2011b, 2012a, 2012b, 2012c) that presents the distributions of the vascular flora of New England in the form of dot distribution maps at the county level (Figure 1). Seven more articles are planned. The atlas is posted on the internet at http://neatlas.org, where it will be updated as new information becomes available. This project encompasses all vascular plants (lycophytes, pteridophytes and spermatophytes) at the rank of species, subspecies, and variety growing independent of cultivation in the six New England states. -
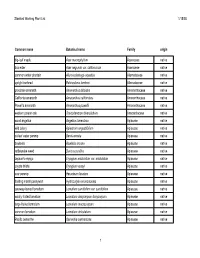
Plant List for Web Page
Stanford Working Plant List 1/15/08 Common name Botanical name Family origin big-leaf maple Acer macrophyllum Aceraceae native box elder Acer negundo var. californicum Aceraceae native common water plantain Alisma plantago-aquatica Alismataceae native upright burhead Echinodorus berteroi Alismataceae native prostrate amaranth Amaranthus blitoides Amaranthaceae native California amaranth Amaranthus californicus Amaranthaceae native Powell's amaranth Amaranthus powellii Amaranthaceae native western poison oak Toxicodendron diversilobum Anacardiaceae native wood angelica Angelica tomentosa Apiaceae native wild celery Apiastrum angustifolium Apiaceae native cutleaf water parsnip Berula erecta Apiaceae native bowlesia Bowlesia incana Apiaceae native rattlesnake weed Daucus pusillus Apiaceae native Jepson's eryngo Eryngium aristulatum var. aristulatum Apiaceae native coyote thistle Eryngium vaseyi Apiaceae native cow parsnip Heracleum lanatum Apiaceae native floating marsh pennywort Hydrocotyle ranunculoides Apiaceae native caraway-leaved lomatium Lomatium caruifolium var. caruifolium Apiaceae native woolly-fruited lomatium Lomatium dasycarpum dasycarpum Apiaceae native large-fruited lomatium Lomatium macrocarpum Apiaceae native common lomatium Lomatium utriculatum Apiaceae native Pacific oenanthe Oenanthe sarmentosa Apiaceae native 1 Stanford Working Plant List 1/15/08 wood sweet cicely Osmorhiza berteroi Apiaceae native mountain sweet cicely Osmorhiza chilensis Apiaceae native Gairdner's yampah (List 4) Perideridia gairdneri gairdneri Apiaceae -

Sinorhizobium Meliloti
Queiroux et al. BMC Microbiology 2012, 12:74 http://www.biomedcentral.com/1471-2180/12/74 RESEARCH ARTICLE Open Access A comparative genomics screen identifies a Sinorhizobium meliloti 1021 sodM-like gene strongly expressed within host plant nodules Clothilde Queiroux1, Brian K Washburn1, Olivia M Davis1,2†, Jamie Stewart1†, Tess E Brewer1, Michael R Lyons1,3 and Kathryn M Jones1* Abstract Background: We have used the genomic data in the Integrated Microbial Genomes system of the Department of Energy’s Joint Genome Institute to make predictions about rhizobial open reading frames that play a role in nodulation of host plants. The genomic data was screened by searching for ORFs conserved in α-proteobacterial rhizobia, but not conserved in closely-related non-nitrogen-fixing α-proteobacteria. Results: Using this approach, we identified many genes known to be involved in nodulation or nitrogen fixation, as well as several new candidate genes. We knocked out selected new genes and assayed for the presence of nodulation phenotypes and/or nodule-specific expression. One of these genes, SMc00911, is strongly expressed by bacterial cells within host plant nodules, but is expressed minimally by free-living bacterial cells. A strain carrying an insertion mutation in SMc00911 is not defective in the symbiosis with host plants, but in contrast to expectations, this mutant strain is able to out-compete the S. meliloti 1021 wild type strain for nodule occupancy in co- inoculation experiments. The SMc00911 ORF is predicted to encode a “SodM-like” (superoxide dismutase-like) protein containing a rhodanese sulfurtransferase domain at the N-terminus and a chromate-resistance superfamily domain at the C-terminus. -

Ensifer (Sinorhizobium) Medicae Strain WSM419
Standards in Genomic Sciences (2010) 2:77-86 DOI:10.4506/sigs.43526 Complete genome sequence of the Medicago microsym- biont Ensifer (Sinorhizobium) medicae strain WSM419 Wayne Reeve1*, Patrick Chain2,3, Graham O’Hara1, Julie Ardley1, Kemanthi Nandesena1, Lambert Bräu1, Ravi Tiwari1, Stephanie Malfatti2,3, Hajnalka Kiss2,3, Alla Lapidus2, Alex Co- peland2, Matt Nolan2, Miriam Land2,4, Loren Hauser2,4, Yun-Juan Chang2,4, Natalia Ivanova2, Konstantinos Mavromatis2, Victor Markowitz5, Nikos Kyrpides2, Margaret Gollagher6, Ron Yates1,7, Michael Dilworth1 & John Howieson1,7. 1 Centre for Rhizobium Studies, Murdoch University, Perth, Australia 2 DOE Joint Genome Institute, Walnut Creek, California, USA 3 Lawrence Livermore National Laboratory, Livermore, California, USA 4 Oak Ridge National Laboratory, Oak Ridge, Tennessee, USA 5 Biological Data Management and Technology Center, Lawrence Berkeley National Labora- tory, Berkeley, California, USA 6 Institute for Sustainability and Technology Policy, Murdoch University, Perth, Australia 7 Department of Agriculture and Food, South Perth, Australia *Corresponding author: Wayne Reeve Keywords: microsymbiont, non-pathogenic, aerobic, Gram-negative rod, root-nodule bacte- ria, nitrogen fixation, Alphaproteobacteria Ensifer (Sinorhizobium) medicae is an effective nitrogen fixing microsymbiont of a diverse range of annual Medicago (medic) species. Strain WSM419 is an aerobic, motile, non-spore forming, Gram-negative rod isolated from a M. murex root nodule collected in Sardinia, Italy in 1981. WSM419 was manufactured commercially in Australia as an inoculant for annual medics during 1985 to 1993 due to its nitrogen fixation, saprophytic competence and acid tolerance properties. Here we describe the basic features of this organism, together with the complete genome sequence, and annotation. This is the first report of a complete genome se- quence for a microsymbiont of the group of annual medic species adapted to acid soils. -

Research Collection
Research Collection Doctoral Thesis Development and application of molecular tools to investigate microbial alkaline phosphatase genes in soil Author(s): Ragot, Sabine A. Publication Date: 2016 Permanent Link: https://doi.org/10.3929/ethz-a-010630685 Rights / License: In Copyright - Non-Commercial Use Permitted This page was generated automatically upon download from the ETH Zurich Research Collection. For more information please consult the Terms of use. ETH Library DISS. ETH NO.23284 DEVELOPMENT AND APPLICATION OF MOLECULAR TOOLS TO INVESTIGATE MICROBIAL ALKALINE PHOSPHATASE GENES IN SOIL A thesis submitted to attain the degree of DOCTOR OF SCIENCES of ETH ZURICH (Dr. sc. ETH Zurich) presented by SABINE ANNE RAGOT Master of Science UZH in Biology born on 25.02.1987 citizen of Fribourg, FR accepted on the recommendation of Prof. Dr. Emmanuel Frossard, examiner PD Dr. Else Katrin Bünemann-König, co-examiner Prof. Dr. Michael Kertesz, co-examiner Dr. Claude Plassard, co-examiner 2016 Sabine Anne Ragot: Development and application of molecular tools to investigate microbial alkaline phosphatase genes in soil, c 2016 ⃝ ABSTRACT Phosphatase enzymes play an important role in soil phosphorus cycling by hydrolyzing organic phosphorus to orthophosphate, which can be taken up by plants and microorgan- isms. PhoD and PhoX alkaline phosphatases and AcpA acid phosphatase are produced by microorganisms in response to phosphorus limitation in the environment. In this thesis, the current knowledge of the prevalence of phoD and phoX in the environment and of their taxonomic distribution was assessed, and new molecular tools were developed to target the phoD and phoX alkaline phosphatase genes in soil microorganisms. -

Wild and Cultivated Clovers of Ohio
WILD AND CULTIVATED CLOVERS OF OHIO. MARY B. LINNELL. FABACEAE—Bean Family. Sub-family—FABATAE. Tribe—Trif olieae—Clovers. Stamens diadelphus, anthers all alike. Leaves with three leaflets, rarely with one leaflet; leaflets denticulate. Synopsis of Genera. I. Corolla falling off after blossoming; petal claws free. 1. Flowers in heads or short racemes, seldom single; pod linear, curved or twisted. a. Pod linear, straight, or somewhat curved, often beaked. Trigonella. b. Pod mostly spirally twisted, sometimes curved, or kidney-shaped. Medicago. 2. Flowers in elongated racemes; pods thick, almost spherical or obovate. Melilotus. II. Corolla mostly drying up and persistent after flowering; petal claws either all or the four lower ones united with the stamen tube. Trifolium. Key. 1. Petals united with the stamen tube, persistent; flowers in globose or elongated heads, or umbellate. Trifolium, 1. Petals free from the stamen tube, falling off. 2. 2. Flowers small, yellow or white, drooping; inflorescence an elongated raceme. Melilotus. 2. Flowers single, in pairs, or in a dense more or less elongated inflorescence.3 3. Leaflets denticulate all around, seldom almost entire-margined; fruit linear, beaked, often somewhat curved. Trigonella. 3. Leaflets denticulate only at the outer end; fruit strongly curved or spirally twisted. Medicago. Trigonella L. Annual plants with yellow or blue flowers. Stipules united with the petiole at the base. Flowers linear, straight or curved. 1. Trigonella foenum-graecum L. Fenugreek. Annual fodder plants; flowers single or in pairs; pod linear, many seeded. Introduced from Asia and cultivated for its aromatic, mucilaginous seeds, formerly employed in medicines and still used by veterinarians. -

Appendix A. Plant Species Known to Occur at Canaveral National Seashore
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science Vegetation Community Monitoring at Canaveral National Seashore, 2009 Natural Resource Data Series NPS/SECN/NRDS—2012/256 ON THE COVER Pitted stripeseed (Piriqueta cistoides ssp. caroliniana) Photograph by Sarah L. Corbett. Vegetation Community Monitoring at Canaveral National Seashore, 2009 Natural Resource Report NPS/SECN/NRDS—2012/256 Michael W. Byrne and Sarah L. Corbett USDI National Park Service Southeast Coast Inventory and Monitoring Network Cumberland Island National Seashore 101 Wheeler Street Saint Marys, Georgia, 31558 and Joseph C. DeVivo USDI National Park Service Southeast Coast Inventory and Monitoring Network University of Georgia 160 Phoenix Road, Phillips Lab Athens, Georgia, 30605 March 2012 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado publishes a range of reports that address natural resource topics of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Data Series is intended for the timely release of basic data sets and data summaries. Care has been taken to assure accuracy of raw data values, but a thorough analysis and interpretation of the data has not been completed. Consequently, the initial analyses of data in this report are provisional and subject to change. All manuscripts in the series receive the appropriate level of peer review to ensure that the information is scientifically credible, technically accurate, appropriately written for the intended audience, and designed and published in a professional manner. -

Les Plantes Hã´Te Des Bruches (Coleoptera Bruchidael : Donnã©Enouvelles Et Corrections
- 277 - Bull. mens. Soc. iinn. Lyon, 2005,74 (7-8) : 277-291 Les plantes hôte des bruches (Coleoptera Bruchidael : donnéenouvelles et corrections. Bernard Delobel* et Alex Delobel- * INWNSA, Laboratoire BF 2 1,20 avenue A. Einstein, F-69621 Villeurbanne cedex ** 47 avenue Paul Langevin, F-92260 Fontenay-aux-Roses Résume- Les auteurs complhtent et corrigent des résultatantérieur (DELOBEL & DELOBEL,2003) sur les relations trophiques entre les bruches de la faune françaiset leurs plan- tes hbtes larvaires. Les nouvelles donnéeconcernent la France, l'Italie et la Grke. Quarante- quatre plantes hr3tes nouvelles ont étidentifiks, et ceci constitue pour plusieurs espècede bru- ches les toutes premihres donnéebiologiques. Le régimalimentaire de 86 % des bruches de ces trois pays est ddsorrnais connu avec une plus ou moins grande prkision. L'hypothès prké deminent emise, selon laquelle chaque espke de broche est infhdé une esee, un genre ou tout au plus à une tribu, et une seule, se trouve confirmde dans la grande majoritÃdes cas. The host plants of seed-beetles (Coleoptera Bruchidae): new data and errata Summary. - The authors complete and modify results published in 2003 on trophic rela- tionships between seed-beetles of the French fauna and their larval host plants. New data are given for France, Italy and Greece. Forty-four new host plants were identified, which constitutes for several beetle species the very tïrsbiological data available. The diet of 86 5% of the se&- batles in the three countries is now more or less precisely known. The previously expressed hypothesis, according to which any given beetle species will feed on a single plant species, genus or tribe, is confimaed in most cases. -

Population Genomics of Sinorhizobium Medicae Based On
Population genomics of Sinorhizobium medicae based on low-coverage sequencing of sympatric isolates Xavier Bailly, Elisa Giuntini, Connor M Sexton, Ryan Pj Lower, Peter W Harrison, Nitin Kumar, J Peter W Young To cite this version: Xavier Bailly, Elisa Giuntini, Connor M Sexton, Ryan Pj Lower, Peter W Harrison, et al.. Popu- lation genomics of Sinorhizobium medicae based on low-coverage sequencing of sympatric isolates. ISME Journal, Nature Publishing Group, 2011, 5 (11), pp.1722-1734. 10.1038/ismej.2011.55. hal- 02652397 HAL Id: hal-02652397 https://hal.inrae.fr/hal-02652397 Submitted on 29 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. The ISME Journal (2011) 5, 1722–1734 & 2011 International Society for Microbial Ecology All rights reserved 1751-7362/11 www.nature.com/ismej ORIGINAL ARTICLE Population genomics of Sinorhizobium medicae based on low-coverage sequencing of sympatric isolates Xavier Bailly1, Elisa Giuntini, M Connor Sexton, Ryan PJ Lower, Peter W Harrison, Nitin Kumar and J Peter W Young Department of Biology, University of York, York, UK We investigated the genomic diversity of a local population of the symbiotic bacterium Sinorhizobium medicae, isolated from the roots of wild Medicago lupulina plants, in order to assess genomic diversity, to identify genomic regions influenced by duplication, deletion or strong selection, and to explore the composition of the pan-genome. -

Norfolk Island Quarantine Survey 2012-2014 – a Comprehensive Assessment of an Isolated Subtropical Island
Norfolk Island Quarantine Survey 2012-2014 – a Comprehensive Assessment of an Isolated Subtropical Island G.V.MAYNARD1, B.J.LEPSCHI2 AND S.F.MALFROY1 1Department of Agriculture and Water Resources, GPO Box 858, Canberra ACT 2601, Australia; and 2Australian National Herbarium, Centre for Australian National Biodiversity Research, GPO Box 1700, Canberra, ACT 2601, Australia Published on 10 March 2018 at https://openjournals.library.sydney.edu.au/index.php/LIN/index Maynard, G.V., Lepschi, B.J. and Malfroy, S.F. (2018). Norfolk Island quarantine survey 2012-2014 – a comprehensive assessment of an isolated subtropical island. Proceedings of the Linnean Society of New South Wales 140, 7-243 A survey of Norfolk Island, Australia was carried out during 2012-2014 to develop a baseline of information on plant pests, and diseases and parasites of domestic animals for biosecurity purposes. The Norfolk Island Quarantine Survey covered introduced vascular plants, invertebrate pests of plants and animals; plant pathogens; pests and diseases of bees, and diseases and parasites of domestic animals. 1747 species were recorded across all organism groups during the course of the survey, of which 658 are newly recorded for Norfolk Island. Details of all organisms recorded during the survey are presented, along with a bibliography of plants and animals of Norfolk Island, with particular reference to introduced taxa. Manuscript received 25 July 2017, accepted for publication 30 January 2018. KEYWORDS: animal diseases, bees, invertebrates, Norfolk Island, plant biosecurity, plant pathogens, plant pests, quarantine survey. INTRODUCTION uninhabited islands - Nepean Island, 1 km to the south, and Philip Island 6 km to the south (Fig. -

NAD1 Controls Defense-Like Responses in Medicago Truncatula Symbiotic Nitrogen Fixing Nodules Following Rhizobial Colonization in a Baca-Independent Manner
G C A T T A C G G C A T genes Article NAD1 Controls Defense-Like Responses in Medicago truncatula Symbiotic Nitrogen Fixing Nodules Following Rhizobial Colonization in a BacA-Independent Manner Ágota Domonkos 1, Szilárd Kovács 2,3, Anikó Gombár 1, Ern˝oKiss 3, Beatrix Horváth 1, Gyöngyi Z. Kováts 1, Attila Farkas 2,Mónika T. Tóth 1, Ferhan Ayaydin 4,Károly Bóka 5, Lili Fodor 1, Pascal Ratet 6,7 ID , Attila Kereszt 2 ID , Gabriella Endre 2,3 and Péter Kaló 1,* 1 National Agricultural and Innovation Center, Agricultural Biotechnology Institute, 2100 Gödöll˝o,Hungary; [email protected] (A.D.); [email protected] (A.G.); [email protected] (B.H.); [email protected] (G.Z.K.); [email protected] (M.T.T.); [email protected] (L.F.); [email protected] (P.K.) 2 Institute of Plant Biology, Biological Research Center, 6726 Szeged, Hungary; [email protected] (S.K.); [email protected] (A.F.); [email protected] (A.K.); [email protected] (G.E.) 3 Institute of Genetics, Biological Research Center, 6726 Szeged, Hungary; [email protected] 4 Cellular Imaging Laboratory, Biological Research Center, 6726 Szeged, Hungary; [email protected] 5 Department of Plant Anatomy, Eötvös Loránd University, 1117 Budapest, Hungary; [email protected] 6 Institute of Plant Sciences Paris-Saclay IPS2, CNRS, INRA, Université Paris-Sud, Université Evry, Université Paris-Saclay, Bâtiment 630, 91405 Orsay, France; [email protected] 7 Institute of Plant Sciences Paris-Saclay IPS2, Paris Diderot, Sorbonne Paris-Cité,Bâtiment 630, 91405 Orsay, France * Correspondence: [email protected]; Tel.: +36-28-526-104 Received: 31 October 2017; Accepted: 11 December 2017; Published: 14 December 2017 Abstract: Legumes form endosymbiotic interaction with host compatible rhizobia, resulting in the development of nitrogen-fixing root nodules.