Table of Contents
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

National Health and Environment Plan
Contents: 1. Respond to health issues posed by pathologies linked to the environment...............12 1.1. Better understanding and prevention of cancers in relations to environmental exposures..................................................................................................................................................... 12 1.1.1 Reduce cancers linked to asbestos..................................................................................13 1.1.2 Take better account of the radon risk in buildings...................................................14 1.2. Prevent health risks linked to exposure to certain plant or animal species.........17 1.3. Better take into account the role of environmental exposures in the increase in certain diseases (metabolic diseases, reproductive diseases, obesity, etc.)..................20 1.3.1 Implement a national strategy on endocrine disrupters........................................20 1.3.2 Understand and act on all environmental factors involved in metabolic diseases and obesity.......................................................................................................................... 21 1.3.3 Prevent reprotoxic risks from environmental exposures and better understand them................................................................................................................................. 23 1.3.4 Prevent neurotoxic risks and neurobehavioural development deficits related to environmental pollutants and better understand them..............................................24 -

120421-24Recombschedule FINAL.Xlsx
Friday 20 April 18:00 20:00 REGISTRATION OPENS in Fira Palace 20:00 21:30 WELCOME RECEPTION in CaixaForum (access map) Saturday 21 April 8:00 8:50 REGISTRATION 8:50 9:00 Opening Remarks (Roderic GUIGÓ and Benny CHOR) Session 1. Chair: Roderic GUIGÓ (CRG, Barcelona ES) 9:00 10:00 Richard DURBIN The Wellcome Trust Sanger Institute, Hinxton UK "Computational analysis of population genome sequencing data" 10:00 10:20 44 Yaw-Ling Lin, Charles Ward and Steven Skiena Synthetic Sequence Design for Signal Location Search 10:20 10:40 62 Kai Song, Jie Ren, Zhiyuan Zhai, Xuemei Liu, Minghua Deng and Fengzhu Sun Alignment-Free Sequence Comparison Based on Next Generation Sequencing Reads 10:40 11:00 178 Yang Li, Hong-Mei Li, Paul Burns, Mark Borodovsky, Gene Robinson and Jian Ma TrueSight: Self-training Algorithm for Splice Junction Detection using RNA-seq 11:00 11:30 coffee break Session 2. Chair: Bonnie BERGER (MIT, Cambrige US) 11:30 11:50 139 Son Pham, Dmitry Antipov, Alexander Sirotkin, Glenn Tesler, Pavel Pevzner and Max Alekseyev PATH-SETS: A Novel Approach for Comprehensive Utilization of Mate-Pairs in Genome Assembly 11:50 12:10 171 Yan Huang, Yin Hu and Jinze Liu A Robust Method for Transcript Quantification with RNA-seq Data 12:10 12:30 120 Zhanyong Wang, Farhad Hormozdiari, Wen-Yun Yang, Eran Halperin and Eleazar Eskin CNVeM: Copy Number Variation detection Using Uncertainty of Read Mapping 12:30 12:50 205 Dmitri Pervouchine Evidence for widespread association of mammalian splicing and conserved long range RNA structures 12:50 13:10 169 Melissa Gymrek, David Golan, Saharon Rosset and Yaniv Erlich lobSTR: A Novel Pipeline for Short Tandem Repeats Profiling in Personal Genomes 13:10 13:30 217 Rory Stark Differential oestrogen receptor binding is associated with clinical outcome in breast cancer 13:30 15:00 lunch break Session 3. -

Methodology for Predicting Semantic Annotations of Protein Sequences by Feature Extraction Derived of Statistical Contact Potentials and Continuous Wavelet Transform
Universidad Nacional de Colombia Sede Manizales Master’s Thesis Methodology for predicting semantic annotations of protein sequences by feature extraction derived of statistical contact potentials and continuous wavelet transform Author: Supervisor: Gustavo Alonso Arango Dr. Cesar German Argoty Castellanos Dominguez A thesis submitted in fulfillment of the requirements for the degree of Master’s on Engineering - Industrial Automation in the Department of Electronic, Electric Engineering and Computation Signal Processing and Recognition Group June 2014 Universidad Nacional de Colombia Sede Manizales Tesis de Maestr´ıa Metodolog´ıapara predecir la anotaci´on sem´antica de prote´ınaspor medio de extracci´on de caracter´ısticas derivadas de potenciales de contacto y transformada wavelet continua Autor: Tutor: Gustavo Alonso Arango Dr. Cesar German Argoty Castellanos Dominguez Tesis presentada en cumplimiento a los requerimientos necesarios para obtener el grado de Maestr´ıaen Ingenier´ıaen Automatizaci´onIndustrial en el Departamento de Ingenier´ıaEl´ectrica,Electr´onicay Computaci´on Grupo de Procesamiento Digital de Senales Enero 2014 UNIVERSIDAD NACIONAL DE COLOMBIA Abstract Faculty of Engineering and Architecture Department of Electronic, Electric Engineering and Computation Master’s on Engineering - Industrial Automation Methodology for predicting semantic annotations of protein sequences by feature extraction derived of statistical contact potentials and continuous wavelet transform by Gustavo Alonso Arango Argoty In this thesis, a method to predict semantic annotations of the proteins from its primary structure is proposed. The main contribution of this thesis lies in the implementation of a novel protein feature representation, which makes use of the pairwise statistical contact potentials describing the protein interactions and geometry at the atomic level. -

Federal Register/Vol. 84, No. 162/Wednesday, August 21, 2019
Federal Register / Vol. 84, No. 162 / Wednesday, August 21, 2019 / Rules and Regulations 43493 Dated: August 15, 2019. 660–0144 or (408) 998–8806 or email Huawei, the ERC determined that there Nazak Nikakhtar, your inquiry to: [email protected]. is reasonable cause to believe that Assistant Secretary of Industry and Analysis, SUPPLEMENTARY INFORMATION: Huawei has been involved in activities International Trade Administration, determined to be contrary to the Performing the Non-Exclusive Duties of the Background national security or foreign policy Under Secretary of Industry and Security. The Entity List (Supplement No. 4 to interests of the United States. In [FR Doc. 2019–17920 Filed 8–19–19; 8:45 am] part 744 of the Export Administration addition, as stated in the May 21 rule, BILLING CODE 3510–33–P Regulations (EAR)) identifies entities for the ERC determined that there was which there is reasonable cause to reasonable cause to believe that the believe, based on specific and affiliates pose a significant risk of DEPARTMENT OF COMMERCE articulable facts, that have been becoming involved in activities contrary involved, are involved, or pose a to the national security or foreign policy Bureau of Industry and Security significant risk of being or becoming interests of the United States due to involved in activities contrary to the their relationship with Huawei. To 15 CFR Part 744 national security or foreign policy illustrate, as set forth in the Superseding interests of the United States. The EAR Indictment filed in the Eastern District [Docket No. 190814–0013] (15 CFR parts 730–774) impose of New York (see the rule published on RIN 0694–AH86 additional license requirements on, and May 21, 2019), Huawei participated limit the availability of all or most along with certain affiliates, including Addition of Certain Entities to the license exceptions for, exports, one or more non-U.S. -

Specific Patterns of Gene Space Organisation Revealed in Wheat by Using the Combination of Barley and Wheat Genomic Resources Camille Rustenholz, Pete E
Specific patterns of gene space organisation revealed in wheat by using the combination of barley and wheat genomic resources Camille Rustenholz, Pete E. Hedley, Jenny Morris, Frédéric Choulet, Catherine Feuillet, Robbie Waugh, Etienne Paux To cite this version: Camille Rustenholz, Pete E. Hedley, Jenny Morris, Frédéric Choulet, Catherine Feuillet, et al.. Specific patterns of gene space organisation revealed in wheat by using the combination of barley and wheat genomic resources. BMC Genomics, BioMed Central, 2010, 11, 10.1186/1471-2164-11-714. hal- 01189739 HAL Id: hal-01189739 https://hal.archives-ouvertes.fr/hal-01189739 Submitted on 30 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Rustenholz et al. BMC Genomics 2010, 11:714 http://www.biomedcentral.com/1471-2164/11/714 RESEARCH ARTICLE Open Access Specific patterns of gene space organisation revealed in wheat by using the combination of barley and wheat genomic resources Camille Rustenholz1†, Pete E Hedley2†, Jenny Morris2, Frédéric Choulet1, Catherine Feuillet1, Robbie Waugh2, Etienne Paux1* Abstract Background: Because of its size, allohexaploid nature and high repeat content, the wheat genome has always been perceived as too complex for efficient molecular studies. -
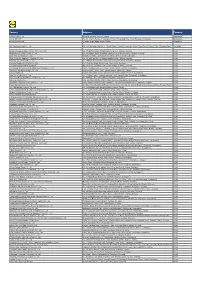
Factory Address Country
Factory Address Country Durable Plastic Ltd. Mulgaon, Kaligonj, Gazipur, Dhaka Bangladesh Lhotse (BD) Ltd. Plot No. 60&61, Sector -3, Karnaphuli Export Processing Zone, North Potenga, Chittagong Bangladesh Bengal Plastics Ltd. Yearpur, Zirabo Bazar, Savar, Dhaka Bangladesh ASF Sporting Goods Co., Ltd. Km 38.5, National Road No. 3, Thlork Village, Chonrok Commune, Korng Pisey District, Konrrg Pisey, Kampong Speu Cambodia Ningbo Zhongyuan Alljoy Fishing Tackle Co., Ltd. No. 416 Binhai Road, Hangzhou Bay New Zone, Ningbo, Zhejiang China Ningbo Energy Power Tools Co., Ltd. No. 50 Dongbei Road, Dongqiao Industrial Zone, Haishu District, Ningbo, Zhejiang China Junhe Pumps Holding Co., Ltd. Wanzhong Villiage, Jishigang Town, Haishu District, Ningbo, Zhejiang China Skybest Electric Appliance (Suzhou) Co., Ltd. No. 18 Hua Hong Street, Suzhou Industrial Park, Suzhou, Jiangsu China Zhejiang Safun Industrial Co., Ltd. No. 7 Mingyuannan Road, Economic Development Zone, Yongkang, Zhejiang China Zhejiang Dingxin Arts&Crafts Co., Ltd. No. 21 Linxian Road, Baishuiyang Town, Linhai, Zhejiang China Zhejiang Natural Outdoor Goods Inc. Xiacao Village, Pingqiao Town, Tiantai County, Taizhou, Zhejiang China Guangdong Xinbao Electrical Appliances Holdings Co., Ltd. South Zhenghe Road, Leliu Town, Shunde District, Foshan, Guangdong China Yangzhou Juli Sports Articles Co., Ltd. Fudong Village, Xiaoji Town, Jiangdu District, Yangzhou, Jiangsu China Eyarn Lighting Ltd. Yaying Gang, Shixi Village, Shishan Town, Nanhai District, Foshan, Guangdong China Lipan Gift & Lighting Co., Ltd. No. 2 Guliao Road 3, Science Industrial Zone, Tangxia Town, Dongguan, Guangdong China Zhan Jiang Kang Nian Rubber Product Co., Ltd. No. 85 Middle Shen Chuan Road, Zhanjiang, Guangdong China Ansen Electronics Co. Ning Tau Administrative District, Qiao Tau Zhen, Dongguan, Guangdong China Changshu Tongrun Auto Accessory Co., Ltd. -

UC San Diego UC San Diego Electronic Theses and Dissertations
UC San Diego UC San Diego Electronic Theses and Dissertations Title Analysis and applications of conserved sequence patterns in proteins Permalink https://escholarship.org/uc/item/5kh9z3fr Author Ie, Tze Way Eugene Publication Date 2007 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, SAN DIEGO Analysis and Applications of Conserved Sequence Patterns in Proteins A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Computer Science by Tze Way Eugene Ie Committee in charge: Professor Yoav Freund, Chair Professor Sanjoy Dasgupta Professor Charles Elkan Professor Terry Gaasterland Professor Philip Papadopoulos Professor Pavel Pevzner 2007 Copyright Tze Way Eugene Ie, 2007 All rights reserved. The dissertation of Tze Way Eugene Ie is ap- proved, and it is acceptable in quality and form for publication on microfilm: Chair University of California, San Diego 2007 iii DEDICATION This dissertation is dedicated in memory of my beloved father, Ie It Sim (1951–1997). iv TABLE OF CONTENTS Signature Page . iii Dedication . iv Table of Contents . v List of Figures . viii List of Tables . x Acknowledgements . xi Vita, Publications, and Fields of Study . xiii Abstract of the Dissertation . xv 1 Introduction . 1 1.1 Protein Homology Search . 1 1.2 Sequence Comparison Methods . 2 1.3 Statistical Analysis of Protein Motifs . 4 1.4 Motif Finding using Random Projections . 5 1.5 Microbial Gene Finding without Assembly . 7 2 Multi-Class Protein Classification using Adaptive Codes . 9 2.1 Profile-based Protein Classifiers . 14 2.2 Embedding Base Classifiers in Code Space . -

Table of Contents
January 12-16, 2019 PLANT & ANIMAL GENOME XXVII Town & Country Hotel THE INTERNATIONAL CONFERENCE San Diego, CA ON THE STATUS OF PLANT & ANIMAL GENOME RESEARCH FINAL PROGRAM & EXHIBIT GUIDE Organizing Committee Chairman: Stephen R. Heller, NIST (USA) Ī PLANT COORGANIZERS Ī ABSTRACTS COORDINATORS Antonio Costa De Oliveira, Federal David Grant, USDA/ARS/CICGR, USA University of Pelotas, Brazil Gerard Lazo, USDA/ARS/WRRC, USA Carolyn J. Lawrence Dill, Iowa State Victoria Carollo Blake, Montana State University, USA University, USA Catherine Feuillet, Inari Agriculture, USA Perry Gustafson, University of Missouri, USA Ī DIGITAL TOOLS AND RESOURCES and University of Nottingham, UK COORDINATORS Fangpu Han, Chinese Academy of Sciences, Monica Munoz-Torres, CGRB, Oregon China State University, USA Brian Smith-White, NIH/NLM/NCBI, USA Jerome P. Miksche, Emeritus Director, USDA, Plant Genome Program, USA TRAVEL GRANTS COORDINATORS Graham Moore, John Innes Centre, UK Ī Tom Blake, Montana State University Rod A. Wing, University of Arizona, USA, (Retired), USA and International Rice Research Institute, Max Rothschild, Iowa State University, USA Philippines Alison L. Van Eenennaam, University of California Davis, USA Ī ANIMAL COORGANIZERS Greger Larson, University of Oxford, UK VISA LETTERS COORDINATOR Molly McCue, University of Minnesota, USA Ī Hans Cheng, USDA/ARS, USA Chris Tuggle, Iowa State University, USA CONFERENCE MANAGEMENT Sponsors and Supporters 184 S. Livingston Avenue Ī USDA, Agricultural Research Service Suite 9, #184 Ī USDA, National Agricultural Library Livingston, New Jersey 07039 USDA, National Institute of Food and Agriculture Ī USA Ī John Innes Centre Phone: (201) 653-5141 Email: [email protected] Cover artwork provided by Applied Biosystems. -

Deep Transcriptome Sequencing Provides New Insights Into The
Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome Lise Pingault, Frédéric Choulet, Adriana Alberti, Natasha Marie Glover, Patrick Wincker, Catherine Feuillet, Etienne Paux To cite this version: Lise Pingault, Frédéric Choulet, Adriana Alberti, Natasha Marie Glover, Patrick Wincker, et al.. Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome. Genome Biology, BioMed Central, 2015, 16 (1), n.p. 10.1186/s13059-015-0601-9. hal-02635440 HAL Id: hal-02635440 https://hal.inrae.fr/hal-02635440 Submitted on 27 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Pingault et al. Genome Biology (2015) 16:29 DOI 10.1186/s13059-015-0601-9 RESEARCH Open Access Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome Lise Pingault1,2, Frédéric Choulet1,2, Adriana Alberti3, Natasha Glover1,2,6, Patrick Wincker3,4,5, Catherine Feuillet1,2,7 and Etienne Paux1,2* Abstract Background: Because of its size, allohexaploid nature, and high repeat content, the bread wheat genome is a good model to study the impact of the genome structure on gene organization, function, and regulation. -

新成立/ 註冊及已更改名稱的公司名單list of Newly Incorporated / Registered Companies and Companies Which Have C
This is the text version of a report with Reference Number "RNC063" and entitled "List of Newly Incorporated /Registered Companies and Companies which have changed Names". The report was created on 01-05-2017 and covers a total of 2828 related records from 24-04-2017 to 30-04-2017. 這是報告編號為「RNC063」,名稱為「新成立 / 註冊及已更改名稱的公司名單」的純文字版報告。這份報告在 2017 年 5 月 1 日建立,包含從 2017 年 4 月 24 日到 2017 年 4 月 30 日到共 2828 個相關紀錄。 Each record in this report is presented in a single row with 6 data fields. Each data field is separated by a "Tab". The order of the 6 data fields are "Sequence Number", "Current Company Name in English", "Current Company Name in Chinese", "C.R. Number", "Date of Incorporation / Registration (D-M-Y)" and "Date of Change of Name (D-M-Y)". 每個紀錄會在報告內被設置成一行,每行細分為 6 個資料。 每個資料會被一個「Tab 符號」分開,6 個資料的次序為「順序編號」、「現用英文公司名稱」、「現用中文公司名稱」、「公司註冊編號」、「成立/註 冊日期(日-月-年)」、「更改名稱日期(日-月-年)」。 Below are the details of records in this report. 以下是這份報告的紀錄詳情。 1. (Hong Kong) Europa Cross - border Electronic Commerce Limited (香港)歐羅巴跨境電子商務有限公司 2529273 24-04-2017 2. 09TypeS XX2000 Limited 2497089 26-04-2017 3. 101 Studio (Eyecare) Limited 2531209 28-04-2017 4. 1VRVR (HK) GROUP LIMITED 醫阿(香港)集團有限公司 2529928 25-04-2017 5. 2Nine Media Limited 2529179 24-04-2017 6. 3FOOD LIMITED 2531104 28-04-2017 7. 3Great Technology Co., Limited 三好科技有限公司 2529287 24-04-2017 8. 4KAAD INTERNATIONAL LIMITED 富凱德國際有限公司 2529819 25-04-2017 9. 777 New Energy Holdings Limited 三七新能源控股有限公司 F0023212 27-04-2017 10. -

The Destruction of Immoral Temples in Qing China Vincent Goossaert
The Destruction of Immoral Temples in Qing China Vincent Goossaert To cite this version: Vincent Goossaert. The Destruction of Immoral Temples in Qing China. ICS Visiting Professor Lectures Series, 2, Chinese University Press, pp.131-153, 2009, Journal of Chinese Studies Special Issue. halshs-00418652 HAL Id: halshs-00418652 https://halshs.archives-ouvertes.fr/halshs-00418652 Submitted on 21 Sep 2009 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. The Destruction of Immoral Temples in Qing China* Vincent Goossaert China, like other pre-modern East Asian countries, was characterized by a pluralistic reli- gious system. Although the Qing Imperial state (1644–1911) recognized this system by granting freedom of practice to a wide range of distinct religious traditions, it was not, for all that matter, tolerant. What was not included within the corpus of officially recognized practices was the object of repression in its various forms. This repression generally remained only a threat, but in some cases the threat was carried out and the reasons and ways in which it was carried out were generally complex and numerous. This article proposes to examine repressive acts as not being necessarily self-evident, as a way of understanding the logic and workings of a state apparatus in its dealings with the religious domain, a particularly difficult area to control. -

SASA Summer 2009 Newsletter
Issue No. 59 Summer 2009 The Fifth Annual Alumni Lecture at SAS Betty Barr April 11-14, 2009, were memorable days for SASA and SAS. On the evening of Saturday, April 11, SAS held its First Annual Gala on the grounds of the YongFoo Elite, at one time the British Consulate, in the former French Con- cession. It was billed as An Evening for the Endowment and will be described more fully in this newsletter by Teddy Heinrichsohn, SASA’s President. That afternoon, however, there had been another significant event: a first informal meeting between alumni of the ‘40s and ‘50s and those of more recent years. Huang Chia Lun, 2000, the organizer, will say more about that. On Sunday, April 12, a group of ten set out in two vans for Jinze, a small town in Qingpu District, about an hour’s drive away from the city, where Deke Erh, has estab- lished a new Research Center. The group comprised Teddy, ’49, Anne Romasco, ’51, Walter Nance, ’50 and Mayna Avent, ‘50, all of whom had flown in to Shanghai a day or so earlier, Dave Merwin, ‘53, who happened to be in Shanghai for a few months studying Chinese, Chia Photo Courtesy of T. Heinrichsohn Lun, Tess Johnston (Old China Hand Research Service), two representatives from the school and me, ‘49. We in SASA were especially happy that the two SAS representatives joined us: Fred Rogers, who will be coming to the school this summer as a new administrator and Rosemary Kinyanjui, the Librarian. Readers of this Newsletter who were at the Salem Reunion in September, 2008, will recall that Deke spent many hours photographing not only the returning alumni but hundreds of their fam- ily photographs which they had been encouraged to bring.