Application-Oriented Marine Isomerases in Biocatalysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Enzymes Handling/Processing
Enzymes Handling/Processing 1 Identification of Petitioned Substance 2 3 This Technical Report addresses enzymes used in used in food processing (handling), which are 4 traditionally derived from various biological sources that include microorganisms (i.e., fungi and 5 bacteria), plants, and animals. Approximately 19 enzyme types are used in organic food processing, from 6 at least 72 different sources (e.g., strains of bacteria) (ETA, 2004). In this Technical Report, information is 7 provided about animal, microbial, and plant-derived enzymes generally, and more detailed information 8 is presented for at least one model enzyme in each group. 9 10 Enzymes Derived from Animal Sources: 11 Commonly used animal-derived enzymes include animal lipase, bovine liver catalase, egg white 12 lysozyme, pancreatin, pepsin, rennet, and trypsin. The model enzyme is rennet. Additional details are 13 also provided for egg white lysozyme. 14 15 Chemical Name: Trade Name: 16 Rennet (animal-derived) Rennet 17 18 Other Names: CAS Number: 19 Bovine rennet 9001-98-3 20 Rennin 25 21 Chymosin 26 Other Codes: 22 Prorennin 27 Enzyme Commission number: 3.4.23.4 23 Rennase 28 24 29 30 31 Chemical Name: CAS Number: 32 Peptidoglycan N-acetylmuramoylhydrolase 9001-63-2 33 34 Other Name: Other Codes: 35 Muramidase Enzyme Commission number: 3.2.1.17 36 37 Trade Name: 38 Egg white lysozyme 39 40 Enzymes Derived from Plant Sources: 41 Commonly used plant-derived enzymes include bromelain, papain, chinitase, plant-derived phytases, and 42 ficin. The model enzyme is bromelain. -

Purification and Properties of Lactate Racemase from Lactobacillus Sake
The Journal of Biochemistry, Vol . 64, No. I, 1968 Purification and Properties of Lactate Racemase from Lactobacillus sake By TETSUG HIYAMA, SAKUzo FUKUI and KAKUO KITAHARA* (From the Institute of Applied Microbiology,University of Tokyo, Bunkyo-ku, Tokyo) (Received for publication, February 12, 1968) A lactate racemase [EC 5.1.2.1] was isolated and purified about 190 fold in specific activity from the sonic extract of Lactobacillus sake. The purified preparation was almost homogeneous by ultracentrifugal and electrophoretical analyses. Characteristic properties of the enzyme were as follows : (a) absorption spectrum showed a single peak at 274 my, (b) molecular weight was 25,000, (c) the enzyme did not require any additional cofactors and showed no activity of lactate dehydrogenase, (d) K, values were 1.7 •~ 10-2 M and 8.0 •~ 10-2 M for D and L-lactate, respectively, (e) optimal pH was 5.8-6.2, (f) an equilibrium point was at a molar ratio of 1/1 (L-isomer/D-isomer), (g) the enzyme activity was inhibited by atebrin, adenosine monosulfate, oxamate and some of Fe chelating agents, (h) pyruvate and acrylate were not incorporated into lactate during the reaction, and (i) exchange reaction of hydrogen between lactate and water did not occur during the reaction. Since the first observation on the enzy the addition of both NAD and pyridoxamine matic racemization of optical active lactate phosphate. In this paper we deal with the by lactic acid bacteria had been reported by purification and properties of the lactate KATAGIRI and KITAHARA in 1936 (1), racemases racemizing enzyme from the cells of L. -
Dr. Martin St. Maurice's Publications
Dr. Martin St. Maurice’s Publications 2013 Lin, Y., and St. Maurice, M. 2013. The structure of allophanate hydrolase from Granulibacter bethesdensis provides insights into substrate specificity in the amidase signature family. Biochemistry. 52: 690-700. 2012 Waldrop, G.L., Holden, H.M., and St. Maurice, M. 2012. The enzymes of biotin dependent CO2 metabolism: What structures reveal about their reaction mechanisms. Protein Science 21(11):1597-1619. Adina-Zada, A., Sereeruk, C., Jitrapakdee, S., Zeczycki, T.N., St. Maurice, M., Cleland, W.W., Wallace, J.C., and Attwood, P.V. 2012. Roles of Arg427 and Arg472 in the binding and allosteric effects of acetyl CoA in pyruvate carboxylase. Biochemistry 51(41): 1597-1619. 2011 Adina-Zada, A., Hazra, R., Sereeruk, C., Jitrapakdee, S., Zeczycki, T.N., St. Maurice, M., Cleland, W.W., Wallace, J.C., and Attwood, P.V. 2011. Probing the allosteric activation of pyruvate carboxylase using 2′,3′-O-(2,4,6-trinitrophenyl) adenosine 5′-triphosphate as a fluorescent mimic of the allosteric activator acetyl CoA. Arch. Biochem. Biophys. 117-126. Zeczycki, T.N., Menefee, A.L., Jitrapakdee, S., Wallace, J.C., Attwood, P.V., St. Maurice, M. and Cleland, W.W. 2011. Activation and inhibition of pyruvate carboxylase from Rhizobium etli. Biochemistry. 9694-9707. Lietzan, A.D., Menefee, A.L., Zeczycki, T.N., Kumar, S., Attwood, P.V., Wallace, J.C., Cleland, W.W. and St. Maurice, M. 2011. Interaction between the biotin carrier domain and the biotin carboxylase domain in the structure of Rhizobium etli pyruvate carboxylase. Biochemistry. 9708-9723. Zeczycki, T.N., Menefee, A.L., Adina-Zada, A., Surinya, K.H., Wallace, J.C., Attwood, P.V., St. -

Table S4. List of Enzymes Directly Involved in the Anti-Oxidant Defense Response
Table S4. List of Enzymes directly involved in the anti-oxidant defense response. Gene Name Gene Symbol Classification/Pathway 6-phosphogluconate dehydrogenase 6PGD NADPH regeneration/Pentose Phosphate Glucose-6-phosphate dehydrogenase G6PD NADPH regeneration/Pentose Phosphate Isocitrate Dehydrogenase 1 IDH1 NADPH regeneration/Krebs Isocitrate Dehydrogenase 2 IDH2 NADPH regeneration/Krebs Malic Enzyme 1 ME1 NADPH regeneration/Krebs Methylenetetrahydrofolate dehydrogenase 1 MTHFD1 NADPH regeneration/Folate Methylenetetrahydrofolate dehydrogenase 2 MTHFD2 NADPH regeneration/Folate Nicotinamide Nucleotide Transhydrogenase NNT NADPH regeneration/NAD Catalase CAT Antioxidants/Catalses/free radical detoxification Glutamate-cysteine ligase catalytic subunit GCLC Antioxidants/Glutathione synthesis Glutamate-cysteine ligase modifier subunit GCLM Antioxidants/Glutathione synthesis Glutathione peroxidase1 GPx1 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase2 GPx2 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase3 GPx3 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase4 GPx4 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase5 GPx5 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase6 GPx6 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase7 GPx7 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione S-transferase -

Regulation of the Tyrosine Kinase Itk by the Peptidyl-Prolyl Isomerase Cyclophilin A
Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A Kristine N. Brazin, Robert J. Mallis, D. Bruce Fulton, and Amy H. Andreotti* Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Ames, IA 50011 Edited by Owen N. Witte, University of California, Los Angeles, CA, and approved December 14, 2001 (received for review October 5, 2001) Interleukin-2 tyrosine kinase (Itk) is a nonreceptor protein tyrosine ulation of the cis and trans conformers. The majority of folded kinase of the Tec family that participates in the intracellular proteins for which three-dimensional structural information has signaling events leading to T cell activation. Tec family members been gathered contain trans prolyl imide bonds. The cis con- contain the conserved SH3, SH2, and catalytic domains common to formation occurs at a frequency of Ϸ6% in folded proteins (17), many kinase families, but they are distinguished by unique se- and a small subset of proteins are conformationally heteroge- quences outside of this region. The mechanism by which Itk and neous with respect to cis͞trans isomerization (18–21). Further- related Tec kinases are regulated is not well understood. Our more, the activation energy for interconversion between cis and studies indicate that Itk catalytic activity is inhibited by the peptidyl trans proline is high (Ϸ20 kcal͞mol) leading to slow intercon- prolyl isomerase activity of cyclophilin A (CypA). NMR structural version rates (22). This barrier is a rate-limiting step in protein studies combined with mutational analysis show that a proline- folding and may serve to kinetically isolate two functionally and dependent conformational switch within the Itk SH2 domain reg- conformationally distinct molecules. -

Pin1 Inhibitors: Towards Understanding the Enzymatic
Pin1 Inhibitors: Towards Understanding the Enzymatic Mechanism Guoyan Xu Dissertation submitted to the faculty of the Virginia Polytechnic Institute and State University In the partial fulfillment of the requirement for the degree of Doctor of Philosophy In Chemistry Felicia A Etzkorn, Chair David G. I. Kingston Neal Castagnoli Paul R. Carlier Brian E. Hanson May 6, 2010 Blacksburg, Virginia Keywords: Pin1, anti-cancer drug target, transition-state analogues, ketoamides, ketones, reduced amides, PPIase assay, inhibition Pin1 Inhibitors: Towards Understanding the Enzymatic Mechanism Guoyan Xu Abstract An important role of Pin1 is to catalyze the cis-trans isomerization of pSer/Thr- Pro bonds; as such, it plays an important role in many cellular events through the effects of conformational change on the function of its biological substrates, including Cdc25, c- Jun, and p53. The expression of Pin1 correlates with cyclin D1 levels, which contributes to cancer cell transformation. Overexpression of Pin1 promotes tumor growth, while its inhibition causes tumor cell apoptosis. Because Pin1 is overexpressed in many human cancer tissues, including breast, prostate, and lung cancer tissues, it plays an important role in oncogenesis, making its study vital for the development of anti-cancer agents. Many inhibitors have been discovered for Pin1, including 1) several classes of designed inhibitors such as alkene isosteres, non-peptidic, small molecular Pin1 inhibitors, and indanyl ketones, and 2) several natural products such as juglone, pepticinnamin E analogues, PiB and its derivatives obtained from a library screen. These Pin1 inhibitors show promise in the development of novel diagnostic and therapeutic anticancer drugs due to their ability to block cell cycle progression. -

Current IUBMB Recommendations on Enzyme Nomenclature and Kinetics$
Perspectives in Science (2014) 1,74–87 Available online at www.sciencedirect.com www.elsevier.com/locate/pisc REVIEW Current IUBMB recommendations on enzyme nomenclature and kinetics$ Athel Cornish-Bowden CNRS-BIP, 31 chemin Joseph-Aiguier, B.P. 71, 13402 Marseille Cedex 20, France Received 9 July 2013; accepted 6 November 2013; Available online 27 March 2014 KEYWORDS Abstract Enzyme kinetics; The International Union of Biochemistry (IUB, now IUBMB) prepared recommendations for Rate of reaction; describing the kinetic behaviour of enzymes in 1981. Despite the more than 30 years that have Enzyme passed since these have not subsequently been revised, though in various respects they do not nomenclature; adequately cover current needs. The IUBMB is also responsible for recommendations on the Enzyme classification naming and classification of enzymes. In contrast to the case of kinetics, these recommenda- tions are kept continuously up to date. & 2014 The Author. Published by Elsevier GmbH. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/3.0/). Contents Introduction...................................................................75 Kinetics introduction...........................................................75 Introduction to enzyme nomenclature ................................................76 Basic definitions ................................................................76 Rates of consumption and formation .................................................76 Rate of reaction .............................................................76 -

Nickel-Pincer Cofactor Biosynthesis Involves Larb-Catalyzed Pyridinium
Nickel-pincer cofactor biosynthesis involves LarB- catalyzed pyridinium carboxylation and LarE- dependent sacrificial sulfur insertion Benoît Desguina,b, Patrice Soumillionb, Pascal Holsb, and Robert P. Hausingera,1 aDepartment of Microbiology and Molecular Genetics, Michigan State University, East Lansing, MI 48824; and bInstitute of Life Sciences, Université catholique de Louvain, B-1348 Louvain-La-Neuve, Belgium Edited by Tadhg P. Begley, Texas A&M University, College Station, TX, and accepted by the Editorial Board March 28, 2016 (received for review January 11, 2016) The lactate racemase enzyme (LarA) of Lactobacillus plantarum proteins) when incubated with ATP (1 mM), MgCl2 (12 mM), harbors a (SCS)Ni(II) pincer complex derived from nicotinic acid. and the other purified proteins, and the activity was stimulated Synthesis of the enzyme-bound cofactor requires LarB, LarC, and fivefold by inclusion of CoA (10 mM) (SI Appendix,Fig.S1A– LarE, which are widely distributed in microorganisms. The functions D). These results are consistent with the requirement of a small of the accessory proteins are unknown, but the LarB C terminus molecule produced by LarB being needed for development of resembles aminoimidazole ribonucleotide carboxylase/mutase, LarC Lar activity. To uncover the substrate of LarB, we replaced the binds Ni and could act in Ni delivery or storage, and LarE is a putative LarB-containing lysates in the LarA activation mixture with ATP-using enzyme of the pyrophosphatase-loop superfamily. Here, purified LarB and potential substrates. Because nicotinic acid is a precursor of the Lar cofactor (1) and LarB features partial we show that LarB carboxylates the pyridinium ring of nicotinic acid sequence identity with PurE, an aminoimidazole ribonucleotide adenine dinucleotide (NaAD) and cleaves the phosphoanhydride (AIR) mutase/carboxylase (4), we tested the possibility that bond to release AMP. -

Letters to Nature
letters to nature Received 7 July; accepted 21 September 1998. 26. Tronrud, D. E. Conjugate-direction minimization: an improved method for the re®nement of macromolecules. Acta Crystallogr. A 48, 912±916 (1992). 1. Dalbey, R. E., Lively, M. O., Bron, S. & van Dijl, J. M. The chemistry and enzymology of the type 1 27. Wolfe, P. B., Wickner, W. & Goodman, J. M. Sequence of the leader peptidase gene of Escherichia coli signal peptidases. Protein Sci. 6, 1129±1138 (1997). and the orientation of leader peptidase in the bacterial envelope. J. Biol. Chem. 258, 12073±12080 2. Kuo, D. W. et al. Escherichia coli leader peptidase: production of an active form lacking a requirement (1983). for detergent and development of peptide substrates. Arch. Biochem. Biophys. 303, 274±280 (1993). 28. Kraulis, P.G. Molscript: a program to produce both detailed and schematic plots of protein structures. 3. Tschantz, W. R. et al. Characterization of a soluble, catalytically active form of Escherichia coli leader J. Appl. Crystallogr. 24, 946±950 (1991). peptidase: requirement of detergent or phospholipid for optimal activity. Biochemistry 34, 3935±3941 29. Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and (1995). the thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281±296 (1991). 4. Allsop, A. E. et al.inAnti-Infectives, Recent Advances in Chemistry and Structure-Activity Relationships 30. Meritt, E. A. & Bacon, D. J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505± (eds Bently, P. H. & O'Hanlon, P. J.) 61±72 (R. Soc. Chem., Cambridge, 1997). -

Fluoride Is a Strong and Specific Inhibitor of (~~Y~~E~R~C~~ Ap,A Hydrolases
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Volume 262, number 2, 205-208 FEBS 08241 March 1990 Fluoride is a strong and specific inhibitor of (~~y~~e~r~c~~ Ap,A hydrolases Andrzej Guranowski Katedra Biackemii, Ak~mia Rolnicza, ul. Wdyirska 35, PL-60-637 Poznati, Pala~d Received 24 November 1989; revised version received 19 January 1990 Fluoride acts as a noncompetitive, strong inhibitor of (usyrmnetricnf) Ap,A hydrolases (EC 361.17). The K, values estimated for the enzymes isolat- ed from seeds of some higher plants (yellow lupin, sunflower and marrow) are in the range of 2-3 pM and Is, for the hydrolase from a mammalian tissue (beef liver) is 20 BM. The anion, up to 25 mM, does not affect the following other enzymes which are able to degrade the bias’-n~l~sidyl~ oligophospha~s: Escherichiu coli (sym~tr~~ AphA hydrolase (EC 3.6.1.41), yeast Ap,A phosphorylsse (EC 2.7.7.53), yellow lupin Ap,A hydrol- ase (EC 3.6.1.29) and phosphodiesterase (EC 3.1.4.1). None of halogenic anions but fluoride affects the activity of (asynrmefricnf) Ap,A hydrolases. Usefulness of the fluoride effect for the in vivo studies on the Ap,A metabolism is shortly discussed. Fluoride; Enzyme inhibition; Ap,A hydrolase 1. INTRODUCTION of the dinucleotides catalyzed by lysyl-, phenylalanyl-, and alanyl-tRNA synthetases [2-51 and inhibits the rat Cellular levels of bis(5 ’ -nucleosidyl)oligophosphates [24] and lupin [l l] Ap3A hydrolases as well as the rat such as Ap.+A, Ap3A or Ap4G are probably modulated liver ApdA hydrolase f25]. -

Exploring the Chemistry and Evolution of the Isomerases
Exploring the chemistry and evolution of the isomerases Sergio Martínez Cuestaa, Syed Asad Rahmana, and Janet M. Thorntona,1 aEuropean Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, United Kingdom Edited by Gregory A. Petsko, Weill Cornell Medical College, New York, NY, and approved January 12, 2016 (received for review May 14, 2015) Isomerization reactions are fundamental in biology, and isomers identifier serves as a bridge between biochemical data and ge- usually differ in their biological role and pharmacological effects. nomic sequences allowing the assignment of enzymatic activity to In this study, we have cataloged the isomerization reactions known genes and proteins in the functional annotation of genomes. to occur in biology using a combination of manual and computa- Isomerases represent one of the six EC classes and are subdivided tional approaches. This method provides a robust basis for compar- into six subclasses, 17 sub-subclasses, and 245 EC numbers cor- A ison and clustering of the reactions into classes. Comparing our responding to around 300 biochemical reactions (Fig. 1 ). results with the Enzyme Commission (EC) classification, the standard Although the catalytic mechanisms of isomerases have already approach to represent enzyme function on the basis of the overall been partially investigated (3, 12, 13), with the flood of new data, an integrated overview of the chemistry of isomerization in bi- chemistry of the catalyzed reaction, expands our understanding of ology is timely. This study combines manual examination of the the biochemistry of isomerization. The grouping of reactions in- chemistry and structures of isomerases with recent developments volving stereoisomerism is straightforward with two distinct types cis-trans in the automatic search and comparison of reactions. -
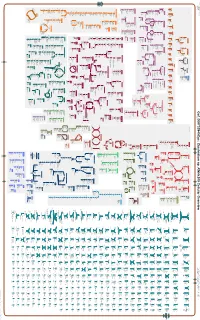
Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Ron Caspi Peter Midford Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000702945Cyc: Clostridium sp. KNHs205 Cellular Overview Connections between pathways are omitted for legibility.