The Genetic Significance of Enzymes in Tissues
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Enzymes Handling/Processing
Enzymes Handling/Processing 1 Identification of Petitioned Substance 2 3 This Technical Report addresses enzymes used in used in food processing (handling), which are 4 traditionally derived from various biological sources that include microorganisms (i.e., fungi and 5 bacteria), plants, and animals. Approximately 19 enzyme types are used in organic food processing, from 6 at least 72 different sources (e.g., strains of bacteria) (ETA, 2004). In this Technical Report, information is 7 provided about animal, microbial, and plant-derived enzymes generally, and more detailed information 8 is presented for at least one model enzyme in each group. 9 10 Enzymes Derived from Animal Sources: 11 Commonly used animal-derived enzymes include animal lipase, bovine liver catalase, egg white 12 lysozyme, pancreatin, pepsin, rennet, and trypsin. The model enzyme is rennet. Additional details are 13 also provided for egg white lysozyme. 14 15 Chemical Name: Trade Name: 16 Rennet (animal-derived) Rennet 17 18 Other Names: CAS Number: 19 Bovine rennet 9001-98-3 20 Rennin 25 21 Chymosin 26 Other Codes: 22 Prorennin 27 Enzyme Commission number: 3.4.23.4 23 Rennase 28 24 29 30 31 Chemical Name: CAS Number: 32 Peptidoglycan N-acetylmuramoylhydrolase 9001-63-2 33 34 Other Name: Other Codes: 35 Muramidase Enzyme Commission number: 3.2.1.17 36 37 Trade Name: 38 Egg white lysozyme 39 40 Enzymes Derived from Plant Sources: 41 Commonly used plant-derived enzymes include bromelain, papain, chinitase, plant-derived phytases, and 42 ficin. The model enzyme is bromelain. -

Tree Kangaroo Conservation Program
Empowered lives. Resilient nations. TREE KANGAROO CONSERVATION PROGRAM Papua New Guinea Equator Initiative Case Studies Local sustainable development solutions for people, nature, and resilient communities UNDP EQUATOR INITIATIVE CASE STUDY SERIES Local and indigenous communities across the world are advancing innovative sustainable development solutions that work for people and for nature. Few publications or case studies tell the full story of how such initiatives evolve, the breadth of their impacts, or how they change over time. Fewer still have undertaken to tell these stories with community practitioners themselves guiding the narrative. The Equator Initiative aims to fill that gap. The Equator Prize 2014 was awarded to 35 outstanding local community and indigenous peoples initiatives working to meet climate and development challenges through the conservation and sustainable use of nature. Selected from 1,234 nomination from across 121 countries, the winners were recognized for their achievements at a prize ceremony held in conjunction with the UN Secretary General’s Climate Summit and the World Conference on Indigenous Peoples in New York City. Special emphasis was placed on forest and ecosystem restoration, food security and agriculture, and water and ocean management. The following case study is one in a growing series that describes vetted and peer-reviewed best practices intended to inspire the policy dialogue needed to take local success to scale, to improve the global knowledge base on local environment and development solutions, and to serve as models for replication. Case studies are best viewed and understood with reference to The Power of Local Action: Lessons from 10 Years of the Equator Prize, a compendium of lessons learned and policy guidance that draws from the case material. -

Table S4. List of Enzymes Directly Involved in the Anti-Oxidant Defense Response
Table S4. List of Enzymes directly involved in the anti-oxidant defense response. Gene Name Gene Symbol Classification/Pathway 6-phosphogluconate dehydrogenase 6PGD NADPH regeneration/Pentose Phosphate Glucose-6-phosphate dehydrogenase G6PD NADPH regeneration/Pentose Phosphate Isocitrate Dehydrogenase 1 IDH1 NADPH regeneration/Krebs Isocitrate Dehydrogenase 2 IDH2 NADPH regeneration/Krebs Malic Enzyme 1 ME1 NADPH regeneration/Krebs Methylenetetrahydrofolate dehydrogenase 1 MTHFD1 NADPH regeneration/Folate Methylenetetrahydrofolate dehydrogenase 2 MTHFD2 NADPH regeneration/Folate Nicotinamide Nucleotide Transhydrogenase NNT NADPH regeneration/NAD Catalase CAT Antioxidants/Catalses/free radical detoxification Glutamate-cysteine ligase catalytic subunit GCLC Antioxidants/Glutathione synthesis Glutamate-cysteine ligase modifier subunit GCLM Antioxidants/Glutathione synthesis Glutathione peroxidase1 GPx1 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase2 GPx2 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase3 GPx3 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase4 GPx4 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase5 GPx5 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase6 GPx6 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione peroxidase7 GPx7 Antioxidants/Glutathione Peroxidases/free radical detoxification Glutathione S-transferase -

Checklist of the Mammals of Indonesia
CHECKLIST OF THE MAMMALS OF INDONESIA Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation i ii CHECKLIST OF THE MAMMALS OF INDONESIA Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation By Ibnu Maryanto Maharadatunkamsi Anang Setiawan Achmadi Sigit Wiantoro Eko Sulistyadi Masaaki Yoneda Agustinus Suyanto Jito Sugardjito RESEARCH CENTER FOR BIOLOGY INDONESIAN INSTITUTE OF SCIENCES (LIPI) iii © 2019 RESEARCH CENTER FOR BIOLOGY, INDONESIAN INSTITUTE OF SCIENCES (LIPI) Cataloging in Publication Data. CHECKLIST OF THE MAMMALS OF INDONESIA: Scientific, English, Indonesia Name and Distribution Area Table in Indonesia Including CITES, IUCN and Indonesian Category for Conservation/ Ibnu Maryanto, Maharadatunkamsi, Anang Setiawan Achmadi, Sigit Wiantoro, Eko Sulistyadi, Masaaki Yoneda, Agustinus Suyanto, & Jito Sugardjito. ix+ 66 pp; 21 x 29,7 cm ISBN: 978-979-579-108-9 1. Checklist of mammals 2. Indonesia Cover Desain : Eko Harsono Photo : I. Maryanto Third Edition : December 2019 Published by: RESEARCH CENTER FOR BIOLOGY, INDONESIAN INSTITUTE OF SCIENCES (LIPI). Jl Raya Jakarta-Bogor, Km 46, Cibinong, Bogor, Jawa Barat 16911 Telp: 021-87907604/87907636; Fax: 021-87907612 Email: [email protected] . iv PREFACE TO THIRD EDITION This book is a third edition of checklist of the Mammals of Indonesia. The new edition provides remarkable information in several ways compare to the first and second editions, the remarks column contain the abbreviation of the specific island distributions, synonym and specific location. Thus, in this edition we are also corrected the distribution of some species including some new additional species in accordance with the discovery of new species in Indonesia. -

A Species-Level Phylogenetic Supertree of Marsupials
J. Zool., Lond. (2004) 264, 11–31 C 2004 The Zoological Society of London Printed in the United Kingdom DOI:10.1017/S0952836904005539 A species-level phylogenetic supertree of marsupials Marcel Cardillo1,2*, Olaf R. P. Bininda-Emonds3, Elizabeth Boakes1,2 and Andy Purvis1 1 Department of Biological Sciences, Imperial College London, Silwood Park, Ascot SL5 7PY, U.K. 2 Institute of Zoology, Zoological Society of London, Regent’s Park, London NW1 4RY, U.K. 3 Lehrstuhl fur¨ Tierzucht, Technical University of Munich, Alte Akademie 12, 85354 Freising-Weihenstephan, Germany (Accepted 26 January 2004) Abstract Comparative studies require information on phylogenetic relationships, but complete species-level phylogenetic trees of large clades are difficult to produce. One solution is to combine algorithmically many small trees into a single, larger supertree. Here we present a virtually complete, species-level phylogeny of the marsupials (Mammalia: Metatheria), built by combining 158 phylogenetic estimates published since 1980, using matrix representation with parsimony. The supertree is well resolved overall (73.7%), although resolution varies across the tree, indicating variation both in the amount of phylogenetic information available for different taxa, and the degree of conflict among phylogenetic estimates. In particular, the supertree shows poor resolution within the American marsupial taxa, reflecting a relative lack of systematic effort compared to the Australasian taxa. There are also important differences in supertrees based on source phylogenies published before 1995 and those published more recently. The supertree can be viewed as a meta-analysis of marsupial phylogenetic studies, and should be useful as a framework for phylogenetically explicit comparative studies of marsupial evolution and ecology. -

Current IUBMB Recommendations on Enzyme Nomenclature and Kinetics$
Perspectives in Science (2014) 1,74–87 Available online at www.sciencedirect.com www.elsevier.com/locate/pisc REVIEW Current IUBMB recommendations on enzyme nomenclature and kinetics$ Athel Cornish-Bowden CNRS-BIP, 31 chemin Joseph-Aiguier, B.P. 71, 13402 Marseille Cedex 20, France Received 9 July 2013; accepted 6 November 2013; Available online 27 March 2014 KEYWORDS Abstract Enzyme kinetics; The International Union of Biochemistry (IUB, now IUBMB) prepared recommendations for Rate of reaction; describing the kinetic behaviour of enzymes in 1981. Despite the more than 30 years that have Enzyme passed since these have not subsequently been revised, though in various respects they do not nomenclature; adequately cover current needs. The IUBMB is also responsible for recommendations on the Enzyme classification naming and classification of enzymes. In contrast to the case of kinetics, these recommenda- tions are kept continuously up to date. & 2014 The Author. Published by Elsevier GmbH. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/3.0/). Contents Introduction...................................................................75 Kinetics introduction...........................................................75 Introduction to enzyme nomenclature ................................................76 Basic definitions ................................................................76 Rates of consumption and formation .................................................76 Rate of reaction .............................................................76 -

Fluoride Is a Strong and Specific Inhibitor of (~~Y~~E~R~C~~ Ap,A Hydrolases
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Volume 262, number 2, 205-208 FEBS 08241 March 1990 Fluoride is a strong and specific inhibitor of (~~y~~e~r~c~~ Ap,A hydrolases Andrzej Guranowski Katedra Biackemii, Ak~mia Rolnicza, ul. Wdyirska 35, PL-60-637 Poznati, Pala~d Received 24 November 1989; revised version received 19 January 1990 Fluoride acts as a noncompetitive, strong inhibitor of (usyrmnetricnf) Ap,A hydrolases (EC 361.17). The K, values estimated for the enzymes isolat- ed from seeds of some higher plants (yellow lupin, sunflower and marrow) are in the range of 2-3 pM and Is, for the hydrolase from a mammalian tissue (beef liver) is 20 BM. The anion, up to 25 mM, does not affect the following other enzymes which are able to degrade the bias’-n~l~sidyl~ oligophospha~s: Escherichiu coli (sym~tr~~ AphA hydrolase (EC 3.6.1.41), yeast Ap,A phosphorylsse (EC 2.7.7.53), yellow lupin Ap,A hydrol- ase (EC 3.6.1.29) and phosphodiesterase (EC 3.1.4.1). None of halogenic anions but fluoride affects the activity of (asynrmefricnf) Ap,A hydrolases. Usefulness of the fluoride effect for the in vivo studies on the Ap,A metabolism is shortly discussed. Fluoride; Enzyme inhibition; Ap,A hydrolase 1. INTRODUCTION of the dinucleotides catalyzed by lysyl-, phenylalanyl-, and alanyl-tRNA synthetases [2-51 and inhibits the rat Cellular levels of bis(5 ’ -nucleosidyl)oligophosphates [24] and lupin [l l] Ap3A hydrolases as well as the rat such as Ap.+A, Ap3A or Ap4G are probably modulated liver ApdA hydrolase f25]. -

Collection of Information on Enzymes a Great Deal of Additional Information on the European Union Is Available on the Internet
European Commission Collection of information on enzymes A great deal of additional information on the European Union is available on the Internet. It can be accessed through the Europa server (http://europa.eu.int). Luxembourg: Office for Official Publications of the European Communities, 2002 ISBN 92-894-4218-2 © European Communities, 2002 Reproduction is authorised provided the source is acknowledged. Final Report „Collection of Information on Enzymes“ Contract No B4-3040/2000/278245/MAR/E2 in co-operation between the Federal Environment Agency Austria Spittelauer Lände 5, A-1090 Vienna, http://www.ubavie.gv.at and the Inter-University Research Center for Technology, Work and Culture (IFF/IFZ) Schlögelgasse 2, A-8010 Graz, http://www.ifz.tu-graz.ac.at PROJECT TEAM (VIENNA / GRAZ) Werner Aberer c Maria Hahn a Manfred Klade b Uli Seebacher b Armin Spök (Co-ordinator Graz) b Karoline Wallner a Helmut Witzani (Co-ordinator Vienna) a a Austrian Federal Environmental Agency (UBA), Vienna b Inter-University Research Center for Technology, Work, and Culture - IFF/IFZ, Graz c University of Graz, Department of Dermatology, Division of Environmental Dermatology, Graz Executive Summary 5 EXECUTIVE SUMMARY Technical Aspects of Enzymes (Chapter 3) Application of enzymes (Section 3.2) Enzymes are applied in various areas of application, the most important ones are technical use, manufacturing of food and feedstuff, cosmetics, medicinal products and as tools for re- search and development. Enzymatic processes - usually carried out under mild conditions - are often replacing steps in traditional chemical processes which were carried out under harsh industrial environments (temperature, pressures, pH, chemicals). Technical enzymes are applied in detergents, for pulp and paper applications, in textile manufacturing, leather industry, for fuel production and for the production of pharmaceuticals and chiral substances in the chemical industry. -

Characterisation, Classification and Conformational Variability Of
Characterisation, Classification and Conformational Variability of Organic Enzyme Cofactors Julia D. Fischer European Bioinformatics Institute Clare Hall College University of Cambridge A thesis submitted for the degree of Doctor of Philosophy 11 April 2011 This dissertation is the result of my own work and includes nothing which is the outcome of work done in collaboration except where specifically indicated in the text. This dissertation does not exceed the word limit of 60,000 words. Acknowledgements I would like to thank all the members of the Thornton research group for their constant interest in my work, their continuous willingness to answer my academic questions, and for their company during my time at the EBI. This includes Saumya Kumar, Sergio Martinez Cuesta, Matthias Ziehm, Dr. Daniela Wieser, Dr. Xun Li, Dr. Irene Pa- patheodorou, Dr. Pedro Ballester, Dr. Abdullah Kahraman, Dr. Rafael Najmanovich, Dr. Tjaart de Beer, Dr. Syed Asad Rahman, Dr. Nicholas Furnham, Dr. Roman Laskowski and Dr. Gemma Holli- day. Special thanks to Asad for allowing me to use early development versions of his SMSD software and for help and advice with the KEGG API installation, to Roman for knowing where to find all kinds of data, to Dani for help with R scripts, to Nick for letting me use his E.C. tree program, to Tjaart for python advice and especially to Gemma for her constant advice and feedback on my work in all aspects, in particular the chemistry side. Most importantly, I would like to thank Prof. Janet Thornton for giving me the chance to work on this project, for all the time she spent in meetings with me and reading my work, for sharing her seemingly limitless knowledge and enthusiasm about the fascinating world of enzymes, and for being such an experienced and motivational advisor. -

On the Evolution of Kangaroos and Their Kin (Family Macropodidae) Using Retrotransposons, Nuclear Genes and Whole Mitochondrial Genomes
ON THE EVOLUTION OF KANGAROOS AND THEIR KIN (FAMILY MACROPODIDAE) USING RETROTRANSPOSONS, NUCLEAR GENES AND WHOLE MITOCHONDRIAL GENOMES William George Dodt B.Sc. (Biochemistry), B.Sc. Hons (Molecular Biology) Principal Supervisor: Dr Matthew J Phillips (EEBS, QUT) Associate Supervisor: Dr Peter Prentis (EEBS, QUT) External Supervisor: Dr Maria Nilsson-Janke (Senckenberg Biodiversity and Research Centre, Frankfurt am Main) Submitted in fulfilment of the requirements for the degree of Doctor of Philosophy Science and Engineering Faculty Queensland University of Technology 2018 1 Keywords Adaptive radiation, ancestral state reconstruction, Australasia, Bayesian inference, endogenous retrovirus, evolution, hybridization, incomplete lineage sorting, incongruence, introgression, kangaroo, Macropodidae, Macropus, mammal, marsupial, maximum likelihood, maximum parsimony, molecular dating, phylogenetics, retrotransposon, speciation, systematics, transposable element 2 Abstract The family Macropodidae contains the kangaroos, wallaroos, wallabies and several closely related taxa that occupy a wide variety of habitats in Australia, New Guinea and surrounding islands. This group of marsupials is the most species rich family within the marsupial order Diprotodontia. Despite significant investigation from previous studies, much of the evolutionary history of macropodids (including their origin within Diprotodontia) has remained unclear, in part due to an incomplete early fossil record. I have utilized several forms of molecular sequence data to shed -
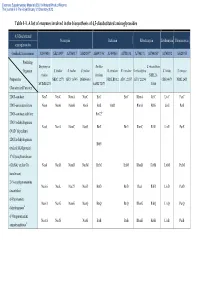
Table 1-1. a List of Enzymes Involved in the Biosynthesis of 4,5-Disubstituted Aminoglycosides
Electronic Supplementary Material (ESI) for Natural Product Reports This journal is © The Royal Society of Chemistry 2013 Table 1-1. A list of enzymes involved in the biosynthesis of 4,5-disubstituted aminoglycosides 4,5-Disubstituted Neomycin Butirosin Ribostamycin Lividomycin Paromomycin aminoglycosides GenBank Accession no. AJ843080 AB211959 AJ786317 AJ629247a AB097196a AJ494863 AJ781030 AJ748131 AJ744850a AJ748832 AJ628955 Producing Streptomyces Bacillus S. ribosidificus Organism S. fradiae S. fradiae S. fradiae B. circulans B. circulans S. ribosidificus S. lividus S. rimosus fradiae circulans NRRL B- Proposed or NBRC 12773 ATCC 10745 DSM 40063 NRRL B3312 ATCC 21557 ATCC 21294 CBS 844.73 NRRL 2455 MCIMB 8233 SANK 72073 11466 Characterized Function 2DOI-synthase Neo7 NeoC NemA NeoC BtrC BtrC RbmA RibC LivC ParC 2DOI-aminotransferase Neo6 NeoB NemB NeoS BtrS BtrR RbmB RibS LivS ParS 2DOI-synthase stabilizer BtrC2b 2DOIA-dehydrogenase Neo5 NeoA NemC NeoE BtrE BtrE RbmC RibE LivE ParE (NAD+ dependent) 2DOIA-dehydrogenase BtrN (radical SAM protein) 1st Glycosyltransferase (GlcNAc and/or Glc Neo8 NeoD NemD NeoM BtrM BtrM RbmD RibM LivM ParM transferase) 2′-N-acetylparomamine Neo16 NeoL Nac25 NeoD BtrD BtrD RacJ RibD LivD ParD deacetylasec 6′-Paromamine Neo11 NeoG NemG NeoQ BtrQ BtrQ RbmG RibQ LivQ ParQ dehydrogenased 6′-Oxoparomamine Neo18 NeoN NeoB BtrB BtrB RbmH RibB LivB ParB aminotransferased Electronic Supplementary Material (ESI) for Natural Product Reports This journal is © The Royal Society of Chemistry 2013 Proposed or Neomycin -

Jessaniniessen2005.Pdf
ARTICLES A streamlined platform for high-content functional proteomics of primary human specimens methods Nadim Jessani1,4,5, Sherry Niessen1,5, BinQing Q Wei1,5, Monica Nicolau2, Mark Humphrey1, Youngran Ji2, Wonshik Han3, Dong-Young Noh3, John R Yates III1, Stefanie S Jeffrey2 & Benjamin F Cravatt1 .com/nature e Achieving information content of satisfactory breadth and depth resolution, facilitating the identification of low-abundance proteins remains a formidable challenge for proteomics. This problem is in complex samples8,9. These techniques, however, are time- .natur w particularly relevant to the study of primary human specimens, consuming (many hours per sample), difficult to perform in such as tumor biopsies, which are heterogeneous and of finite parallel and require large quantities of proteome (B1mgor quantity. Here we present a functional proteomics strategy that greater) for optimal analysis. Alternative proteomic approaches unites the activity-based protein profiling and multidimensional http://ww have emerged that have greater throughput (matrix-assisted laser protein identification technologies (ABPP-MudPIT) for the desorption ionization (MALDI) imaging6 and surface-enhanced 7 oup streamlined analysis of human samples. This convergent laser desorption ionization time-of-flight (SELDI-TOF) spectro- r G platform involves a rapid initial phase, in which enzyme activity metry), albeit at the expense of sensitivity. Furthermore, each of signatures are generated for functional classification of samples, these methods, by focusing on measurements of protein expression, followed by in-depth analysis of representative members from provides only an indirect readout of protein activity, which is lishing each class. Using this two-tiered approach, we identified more regulated by a myriad of post-translational events in vivo10.