Original Article Bioinformatic Analysis of Gene Expression Profile in Prostate Epithelial Cells Exposed to Low-Dose Cadmium
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dual Mutations Reveal Interactions Between Components of Oxidative Phosphorylation in Kluyveromyces Lactis
Copyright 2001 by the Genetics Society of America Dual Mutations Reveal Interactions Between Components of Oxidative Phosphorylation in Kluyveromyces lactis G. D. Clark-Walker and X. J. Chen Molecular Genetics and Evolution Group, Research School of Biological Sciences, The Australian National University, Canberra, ACT, 2601, Australia Manuscript received April 10, 2001 Accepted for publication August 3, 2001 ABSTRACT Loss of mtDNA or mitochondrial protein synthesis cannot be tolerated by wild-type Kluyveromyces lactis. The mitochondrial function responsible for 0-lethality has been identified by disruption of nuclear genes encoding electron transport and F0-ATP synthase components of oxidative phosphorylation. Sporulation of diploid strains heterozygous for disruptions in genes for the two components of oxidative phosphoryla- tion results in the formation of nonviable spores inferred to contain both disruptions. Lethality of spores is thought to result from absence of a transmembrane potential, ⌬⌿, across the mitochondrial inner membrane due to lack of proton pumping by the electron transport chain or reversal of F1F0-ATP synthase. Synergistic lethality, caused by disruption of nuclear genes, or 0-lethality can be suppressed by the atp2.1  mutation in the -subunit of F1-ATPase. Suppression is viewed as occurring by an increased hydrolysis of ATP by mutant F1, allowing sufficient electrogenic exchange by the translocase of ADP in the matrix for ATP in the cytosol to maintain ⌬⌿. In addition, lethality of haploid strains with a disruption of AAC encoding the ADP/ATP translocase can be suppressed by atp2.1. In this case suppression is considered to occur by mutant F1 acting in the forward direction to partially uncouple ATP production, thereby stimulating respiration and relieving detrimental hyperpolarization of the inner membrane. -

Low Abundance of the Matrix Arm of Complex I in Mitochondria Predicts Longevity in Mice
ARTICLE Received 24 Jan 2014 | Accepted 9 Apr 2014 | Published 12 May 2014 DOI: 10.1038/ncomms4837 OPEN Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice Satomi Miwa1, Howsun Jow2, Karen Baty3, Amy Johnson1, Rafal Czapiewski1, Gabriele Saretzki1, Achim Treumann3 & Thomas von Zglinicki1 Mitochondrial function is an important determinant of the ageing process; however, the mitochondrial properties that enable longevity are not well understood. Here we show that optimal assembly of mitochondrial complex I predicts longevity in mice. Using an unbiased high-coverage high-confidence approach, we demonstrate that electron transport chain proteins, especially the matrix arm subunits of complex I, are decreased in young long-living mice, which is associated with improved complex I assembly, higher complex I-linked state 3 oxygen consumption rates and decreased superoxide production, whereas the opposite is seen in old mice. Disruption of complex I assembly reduces oxidative metabolism with concomitant increase in mitochondrial superoxide production. This is rescued by knockdown of the mitochondrial chaperone, prohibitin. Disrupted complex I assembly causes premature senescence in primary cells. We propose that lower abundance of free catalytic complex I components supports complex I assembly, efficacy of substrate utilization and minimal ROS production, enabling enhanced longevity. 1 Institute for Ageing and Health, Newcastle University, Newcastle upon Tyne NE4 5PL, UK. 2 Centre for Integrated Systems Biology of Ageing and Nutrition, Newcastle University, Newcastle upon Tyne NE4 5PL, UK. 3 Newcastle University Protein and Proteome Analysis, Devonshire Building, Devonshire Terrace, Newcastle upon Tyne NE1 7RU, UK. Correspondence and requests for materials should be addressed to T.v.Z. -

(BPA) Exposure Biomarkers in Ovarian Cancer
Journal of Clinical Medicine Article Identification of Potential Bisphenol A (BPA) Exposure Biomarkers in Ovarian Cancer Aeman Zahra 1, Qiduo Dong 1, Marcia Hall 1,2 , Jeyarooban Jeyaneethi 1, Elisabete Silva 1, Emmanouil Karteris 1,* and Cristina Sisu 1,* 1 Biosciences, College of Health, Medicine and Life Sciences, Brunel University London, Uxbridge UB8 3PH, UK; [email protected] (A.Z.); [email protected] (Q.D.); [email protected] (M.H.); [email protected] (J.J.); [email protected] (E.S.) 2 Mount Vernon Cancer Centre, Northwood HA6 2RN, UK * Correspondence: [email protected] (E.K.); [email protected] (C.S.) Abstract: Endocrine-disrupting chemicals (EDCs) can exert multiple deleterious effects and have been implicated in carcinogenesis. The xenoestrogen Bisphenol A (BPA) that is found in various consumer products has been involved in the dysregulation of numerous signalling pathways. In this paper, we present the analysis of a set of 94 genes that have been shown to be dysregulated in presence of BPA in ovarian cancer cell lines since we hypothesised that these genes might be of biomarker potential. This study sought to identify biomarkers of disease and biomarkers of disease- associated exposure. In silico analyses took place using gene expression data extracted from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) databases. Differential expression was further validated at protein level using immunohistochemistry on an ovarian cancer tissue microarray. We found that 14 out of 94 genes are solely dysregulated in the presence of BPA, while the remaining 80 genes are already dysregulated (p-value < 0.05) in their expression pattern Citation: Zahra, A.; Dong, Q.; Hall, as a consequence of the disease. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Molecular and Biochemical Characterisation of the Electron Transport Chain of Plasmodium Falciparum
Molecular and biochemical characterisation of the electron transport chain of Plasmodium falciparum Thomas ANTOINE Ph.D. February 2012 Molecular and biochemical characterisation of the electron transport chain of Plasmodium falciparum Thesis is submitted in accordance with the requirements of the University of Liverpool for the degree of Doctor of Philosophy By Thomas ANTOINE February 2012 Declaration This thesis is the result of my own work. The material contained in the thesis has not been presented, nor is currently being presented, either wholly or as a part, for any other degree or other qualification. The research work was carried out in the Liverpool School of Tropical Medicine, University of Liverpool, United Kingdom. ……………………………………….. Thomas ANTOINE (2012) i Acknowledgments First of all, I would like to express my gratitude to my supervisors Pr. Steve Ward, Dr. Giancarlo Biagini and Dr. Nick Fisher for their supervision, continued support and constant encouragement throughout my PhD. A special thank goes to the InterMal PhD program and the Marie Curie Fellowship for providing the generous funding which allowed me to undertake this research, but also for giving me the opportunity to attend conferences and workshops as well as meet so many interesting people. I am grateful to Prof. Alister Craig and Prof. Sylke Muller for accepting to be the two members of my oral defense committee. The members of the Ward's research group have contributed immensely to my personal and professional time at Liverpool. I am very grateful to Ashley Warman for insightful comments in my work, for reviewing my thesis and providing valuable feedback as well as for many motivating discussions. -

THE FUNCTIONAL SIGNIFICANCE of MITOCHONDRIAL SUPERCOMPLEXES in C. ELEGANS by WICHIT SUTHAMMARAK Submitted in Partial Fulfillment
THE FUNCTIONAL SIGNIFICANCE OF MITOCHONDRIAL SUPERCOMPLEXES in C. ELEGANS by WICHIT SUTHAMMARAK Submitted in partial fulfillment of the requirements For the degree of Doctor of Philosophy Dissertation Advisor: Drs. Margaret M. Sedensky & Philip G. Morgan Department of Genetics CASE WESTERN RESERVE UNIVERSITY January, 2011 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of _____________________________________________________ candidate for the ______________________degree *. (signed)_______________________________________________ (chair of the committee) ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. Dedicated to my family, my teachers and all of my beloved ones for their love and support ii ACKNOWLEDGEMENTS My advanced academic journey began 5 years ago on the opposite side of the world. I traveled to the United States from Thailand in search of a better understanding of science so that one day I can return to my homeland and apply the knowledge and experience I have gained to improve the lives of those affected by sickness and disease yet unanswered by science. Ultimately, I hoped to make the academic transition into the scholarly community by proving myself through scientific research and understanding so that I can make a meaningful contribution to both the scientific and medical communities. The following dissertation would not have been possible without the help, support, and guidance of a lot of people both near and far. I wish to thank all who have aided me in one way or another on this long yet rewarding journey. My sincerest thanks and appreciation goes to my advisors Philip Morgan and Margaret Sedensky. -

Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces Cerevisiae
life Review Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae Leticia V. R. Franco 1,2,* , Luca Bremner 1 and Mario H. Barros 2 1 Department of Biological Sciences, Columbia University, New York, NY 10027, USA; [email protected] 2 Department of Microbiology,Institute of Biomedical Sciences, Universidade de Sao Paulo, Sao Paulo 05508-900, Brazil; [email protected] * Correspondence: [email protected] Received: 27 October 2020; Accepted: 19 November 2020; Published: 23 November 2020 Abstract: The ease with which the unicellular yeast Saccharomyces cerevisiae can be manipulated genetically and biochemically has established this organism as a good model for the study of human mitochondrial diseases. The combined use of biochemical and molecular genetic tools has been instrumental in elucidating the functions of numerous yeast nuclear gene products with human homologs that affect a large number of metabolic and biological processes, including those housed in mitochondria. These include structural and catalytic subunits of enzymes and protein factors that impinge on the biogenesis of the respiratory chain. This article will review what is currently known about the genetics and clinical phenotypes of mitochondrial diseases of the respiratory chain and ATP synthase, with special emphasis on the contribution of information gained from pet mutants with mutations in nuclear genes that impair mitochondrial respiration. Our intent is to provide the yeast mitochondrial specialist with basic knowledge of human mitochondrial pathologies and the human specialist with information on how genes that directly and indirectly affect respiration were identified and characterized in yeast. Keywords: mitochondrial diseases; respiratory chain; yeast; Saccharomyces cerevisiae; pet mutants 1. -

Snps) in Genes of the Alcohol Dehydrogenase, Glutathione S-Transferase, and Nicotinamide Adenine Dinucleotide, Reduced (NADH) Ubiquinone Oxidoreductase Families
J Hum Genet (2001) 46:385–407 © Jpn Soc Hum Genet and Springer-Verlag 2001 ORIGINAL ARTICLE Aritoshi Iida · Susumu Saito · Akihiro Sekine Takuya Kitamoto · Yuri Kitamura · Chihiro Mishima Saori Osawa · Kimie Kondo · Satoko Harigae Yusuke Nakamura Catalog of 434 single-nucleotide polymorphisms (SNPs) in genes of the alcohol dehydrogenase, glutathione S-transferase, and nicotinamide adenine dinucleotide, reduced (NADH) ubiquinone oxidoreductase families Received: March 15, 2001 / Accepted: April 6, 2001 Abstract An approach based on development of a large Introduction archive of single-nucleotide polymorphisms (SNPs) throughout the human genome is expected to facilitate large-scale studies to identify genes associated with drug Human genetic variations result from a dynamic process efficacy and side effects, or susceptibility to common over time that includes sudden mutations, random genetic diseases. We have already described collections of SNPs drift, and, in some cases, founder effects. Variations at a present among various genes encoding drug-metabolizing single gene locus are useful as markers of individual risk for enzymes. Here we report SNPs for such enzymes at adverse drug reactions or susceptibility to complex diseases additional loci, including 8 alcohol dehydrogenases, 12 (for reviews, see Risch and Merikangas 1996; Kruglyak glutathione S-transferases, and 18 belonging to the NADH- 1997; McCarthy and Hilfiker 2000). Common types of ubiquinone oxidoreductase family. Among DNA samples sequence variation in the human genome include single- from 48 Japanese volunteers, we identified a total of 434 nucleotide polymorphisms (SNPs), insertion/deletion SNPs at these 38 loci: 27 within coding elements, 52 in 59 polymorphisms, and variations in the number of repeats of flanking regions, five in 59 untranslated regions, 293 in certain motifs (e.g., microsatellites and variable number of introns, 20 in 39 untranslated regions, and 37 in 39 flanking tandem repeat loci). -
Download Validation Data
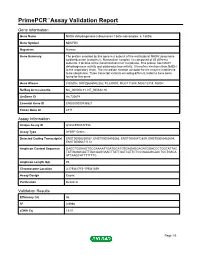
PrimePCR™Assay Validation Report Gene Information Gene Name NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa Gene Symbol NDUFB5 Organism Human Gene Summary The protein encoded by this gene is a subunit of the multisubunit NADH:ubiquinone oxidoreductase (complex I). Mammalian complex I is composed of 45 different subunits. It locates at the mitochondrial inner membrane. This protein has NADH dehydrogenase activity and oxidoreductase activity. It transfers electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone. Three transcript variants encoding different isoforms have been found for this gene. Gene Aliases CISGDH, DKFZp686N02262, FLJ30597, MGC111204, MGC12314, SGDH RefSeq Accession No. NC_000003.11, NT_005612.16 UniGene ID Hs.730674 Ensembl Gene ID ENSG00000136521 Entrez Gene ID 4711 Assay Information Unique Assay ID qHsaCED0037586 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000259037, ENST00000493866, ENST00000472629, ENST00000482604, ENST00000471112 Amplicon Context Sequence GAGCTGGAAGTGCGAAAATTGATGCATGTGAGAGGAGATGGACCCTGGTATTAC TATGAGACAATTGACAAGGAACTTATTGATCATTCTCCGAAAGCAACTCCTGACA ATTAAGCATTTTTTTC Amplicon Length (bp) 95 Chromosome Location 3:179341715-179341839 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 96 R2 0.9998 cDNA Cq 19.01 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 79 gDNA Cq 24.09 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. -

Electron Transport Chain Activity Is a Predictor and Target for Venetoclax Sensitivity in Multiple Myeloma
ARTICLE https://doi.org/10.1038/s41467-020-15051-z OPEN Electron transport chain activity is a predictor and target for venetoclax sensitivity in multiple myeloma Richa Bajpai1,7, Aditi Sharma 1,7, Abhinav Achreja2,3, Claudia L. Edgar1, Changyong Wei1, Arusha A. Siddiqa1, Vikas A. Gupta1, Shannon M. Matulis1, Samuel K. McBrayer 4, Anjali Mittal3,5, Manali Rupji 6, Benjamin G. Barwick 1, Sagar Lonial1, Ajay K. Nooka 1, Lawrence H. Boise 1, Deepak Nagrath2,3,5 & ✉ Mala Shanmugam 1 1234567890():,; The BCL-2 antagonist venetoclax is highly effective in multiple myeloma (MM) patients exhibiting the 11;14 translocation, the mechanistic basis of which is unknown. In evaluating cellular energetics and metabolism of t(11;14) and non-t(11;14) MM, we determine that venetoclax-sensitive myeloma has reduced mitochondrial respiration. Consistent with this, low electron transport chain (ETC) Complex I and Complex II activities correlate with venetoclax sensitivity. Inhibition of Complex I, using IACS-010759, an orally bioavailable Complex I inhibitor in clinical trials, as well as succinate ubiquinone reductase (SQR) activity of Complex II, using thenoyltrifluoroacetone (TTFA) or introduction of SDHC R72C mutant, independently sensitize resistant MM to venetoclax. We demonstrate that ETC inhibition increases BCL-2 dependence and the ‘primed’ state via the ATF4-BIM/NOXA axis. Further, SQR activity correlates with venetoclax sensitivity in patient samples irrespective of t(11;14) status. Use of SQR activity in a functional-biomarker informed manner may better select for MM patients responsive to venetoclax therapy. 1 Department of Hematology and Medical Oncology, Winship Cancer Institute, School of Medicine, Emory University, Atlanta, GA, USA. -

A High-Fat Diet Coordinately Downregulates Genes Required for Mitochondrial Oxidative Phosphorylation in Skeletal Muscle Lauren M
A High-Fat Diet Coordinately Downregulates Genes Required for Mitochondrial Oxidative Phosphorylation in Skeletal Muscle Lauren M. Sparks, Hui Xie, Robert A. Koza, Randall Mynatt, Matthew W. Hulver, George A. Bray, and Steven R. Smith Obesity and type 2 diabetes have been associated with a ray studies have shown that genes involved in oxidative high-fat diet (HFD) and reduced mitochondrial mass phosphorylation (OXPHOS) exhibit reduced expression and function. We hypothesized a HFD may affect expres- levels in the skeletal muscle of type 2 diabetic subjects and sion of genes involved in mitochondrial function and prediabetic subjects. These changes may be mediated by biogenesis. To test this hypothesis, we fed 10 insulin- the peroxisome proliferator–activated receptor ␥ coacti- sensitive males an isoenergetic HFD for 3 days with vator-1 (PGC1) pathway. PGC1␣- and PGC1-responsive muscle biopsies before and after intervention. Oligonu- cleotide microarray analysis revealed 297 genes were OXPHOS genes show reduced expression in the muscle of differentially regulated by the HFD (Bonferonni ad- patients with type 2 diabetes (3,4). In addition to the justed P < 0.001). Six genes involved in oxidative cellular energy sensor AMP kinase, the peroxisome prolif- phosphorylation (OXPHOS) decreased. Four were mem- erator–activated receptor cofactors PGC1␣ (5–7) and pos- bers of mitochondrial complex I: NDUFB3, NDUFB5, sibly PGC1 (8) activate mitochondrial biogenesis and NDUFS1, and NDUFV1; one was SDHB in complex II and increase OXPHOS gene expression by increasing the tran- a mitochondrial carrier protein SLC25A12. Peroxisome scription, translation, and activation of the transcription proliferator–activated receptor ␥ coactivator-1 (PGC1) -and PGC1 mRNA were decreased by ؊20%, P < 0.01, factors necessary for mitochondrial DNA (mtDNA) repli ␣ and ؊25%, P < 0.01, respectively. -

Mouse Ndufb5 Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Ndufb5 Knockout Project (CRISPR/Cas9) Objective: To create a Ndufb5 knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Ndufb5 gene (NCBI Reference Sequence: NM_025316 ; Ensembl: ENSMUSG00000027673 ) is located on Mouse chromosome 3. 6 exons are identified, with the ATG start codon in exon 1 and the TGA stop codon in exon 6 (Transcript: ENSMUST00000127477). Exon 2~5 will be selected as target site. Cas9 and gRNA will be co-injected into fertilized eggs for KO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Exon 2 starts from about 22.05% of the coding region. Exon 2~5 covers 57.32% of the coding region. The size of effective KO region: ~3714 bp. The KO region does not have any other known gene. Page 1 of 9 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 2 3 4 5 6 Legends Exon of mouse Ndufb5 Knockout region Page 2 of 9 https://www.alphaknockout.com Overview of the Dot Plot (up) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section upstream of Exon 2 is aligned with itself to determine if there are tandem repeats. No significant tandem repeat is found in the dot plot matrix. So this region is suitable for PCR screening or sequencing analysis. Overview of the Dot Plot (down) Window size: 15 bp Forward Reverse Complement Sequence 12 Note: The 2000 bp section downstream of Exon 5 is aligned with itself to determine if there are tandem repeats.