Coordinated Regulatory Variation Associated with Gestational Hyperglycaemia Regulates Expression of the Novel Hexokinase HKDC1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ck1δ Over-Expressing Mice Display ADHD-Like Behaviors, Frontostriatal Neuronal Abnormalities and Altered Expressions of ADHD-Candidate Genes
Molecular Psychiatry (2020) 25:3322–3336 https://doi.org/10.1038/s41380-018-0233-z ARTICLE CK1δ over-expressing mice display ADHD-like behaviors, frontostriatal neuronal abnormalities and altered expressions of ADHD-candidate genes 1 1 1 2 1 1 1 Mingming Zhou ● Jodi Gresack ● Jia Cheng ● Kunihiro Uryu ● Lars Brichta ● Paul Greengard ● Marc Flajolet Received: 8 November 2017 / Revised: 4 July 2018 / Accepted: 18 July 2018 / Published online: 19 October 2018 © Springer Nature Limited 2018 Abstract The cognitive mechanisms underlying attention-deficit hyperactivity disorder (ADHD), a highly heritable disorder with an array of candidate genes and unclear genetic architecture, remain poorly understood. We previously demonstrated that mice overexpressing CK1δ (CK1δ OE) in the forebrain show hyperactivity and ADHD-like pharmacological responses to D- amphetamine. Here, we demonstrate that CK1δ OE mice exhibit impaired visual attention and a lack of D-amphetamine- induced place preference, indicating a disruption of the dopamine-dependent reward pathway. We also demonstrate the presence of abnormalities in the frontostriatal circuitry, differences in synaptic ultra-structures by electron microscopy, as 1234567890();,: 1234567890();,: well as electrophysiological perturbations of both glutamatergic and GABAergic transmission, as observed by altered frequency and amplitude of mEPSCs and mIPSCs. Furthermore, gene expression profiling by next-generation sequencing alone, or in combination with bacTRAP technology to study specifically Drd1a versus Drd2 medium spiny neurons, revealed that developmental CK1δ OE alters transcriptional homeostasis in the striatum, including specific alterations in Drd1a versus Drd2 neurons. These results led us to perform a fine molecular characterization of targeted gene networks and pathway analysis. Importantly, a large fraction of 92 genes identified by GWAS studies as associated with ADHD in humans are significantly altered in our mouse model. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Supplementary Table 1. List of Genes Up-Regulated in Abiraterone-Resistant Vcap Xenograft Samples PIK3IP1 Phosphoinositide-3-Kin
Supplementary Table 1. List of genes up-regulated in abiraterone-resistant VCaP xenograft samples PIK3IP1 phosphoinositide-3-kinase interacting protein 1 TMEM45A transmembrane protein 45A THBS1 thrombospondin 1 C7orf63 chromosome 7 open reading frame 63 OPTN optineurin FAM49A family with sequence similarity 49, member A APOL4 apolipoprotein L, 4 C17orf108|LOC201229 chromosome 17 open reading frame 108 | hypothetical protein LOC201229 SNORD94 small nucleolar RNA, C/D box 94 PCDHB11 protocadherin beta 11 RBM11 RNA binding motif protein 11 C6orf225 chromosome 6 open reading frame 225 KIAA1984|C9orf86|TMEM14 1 KIAA1984 | chromosome 9 open reading frame 86 | transmembrane protein 141 KIAA1107 TLR3 toll-like receptor 3 LPAR6 lysophosphatidic acid receptor 6 KIAA1683 GRB10 growth factor receptor-bound protein 10 TIMP2 TIMP metallopeptidase inhibitor 2 CCDC28A coiled-coil domain containing 28A FBXL2 F-box and leucine-rich repeat protein 2 NOV nephroblastoma overexpressed gene TSPAN31 tetraspanin 31 NR3C2 nuclear receptor subfamily 3, group C, member 2 DYNC2LI1 dynein, cytoplasmic 2, light intermediate chain 1 C15orf51 dynamin 1 pseudogene SAMD13 sterile alpha motif domain containing 13 RASSF6 Ras association (RalGDS/AF-6) domain family member 6 ZNF167 zinc finger protein 167 GATA2 GATA binding protein 2 NUDT7 nudix (nucleoside diphosphate linked moiety X)-type motif 7 DNAJC18 DnaJ (Hsp40) homolog, subfamily C, member 18 SNORA57 small nucleolar RNA, H/ACA box 57 CALCOCO1 calcium binding and coiled-coil domain 1 RLN2 relaxin 2 ING4 inhibitor of -

A Curated Biochemical Pathways Database for the Laboratory Mouse Alexei V Evsikov, Mary E Dolan, Michael P Genrich, Emily Patek and Carol J Bult
Open Access Software2009EvsikovetVolume al. 10, Issue 8, Article R84 MouseCyc: a curated biochemical pathways database for the laboratory mouse Alexei V Evsikov, Mary E Dolan, Michael P Genrich, Emily Patek and Carol J Bult Address: The Jackson Laboratory, Main Street, Bar Harbor, ME 04609, USA. Correspondence: Carol J Bult. Email: [email protected] Published: 14 August 2009 Received: 22 May 2009 Revised: 17 July 2009 Genome Biology 2009, 10:R84 (doi:10.1186/gb-2009-10-8-r84) Accepted: 14 August 2009 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2009/10/8/R84 © 2009 Evsikov et al,; licensee BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. MouseCyc<p>MouseCyc database is a database of curated metabolic pathways for the laboratory mouse.</p> Abstract Linking biochemical genetic data to the reference genome for the laboratory mouse is important for comparative physiology and for developing mouse models of human biology and disease. We describe here a new database of curated metabolic pathways for the laboratory mouse called MouseCyc http://mousecyc.jax.org. MouseCyc has been integrated with genetic and genomic data for the laboratory mouse available from the Mouse Genome Informatics database and with pathway data from other organisms, including human. Rationale Biochemical interactions and transformations among organic The availability of the nearly complete genome sequence for molecules are arguably the foundation and core distinguish- the laboratory mouse provides a powerful platform for pre- ing feature of all organic life. -

Molecular Evolution of the Vertebrate Hexokinase Gene Family: Identification of a Conserved Fifth Vertebrate Hexokinase Gene ⁎ David M
中国科技论文在线 http://www.paper.edu.cn Comparative Biochemistry and Physiology, Part D 3 (2008) 96–107 Molecular evolution of the vertebrate hexokinase gene family: Identification of a conserved fifth vertebrate hexokinase gene ⁎ David M. Irwin a, , Huanran Tan b a Department of Laboratory Medicine and Pathobiology, Banting and Best Diabetes Centre, University of Toronto, Toronto, Canada b Department of Pharmacology, Peking University Health Sciences Centre, Beijing, China Received 6 October 2007; received in revised form 12 November 2007; accepted 13 November 2007 Available online 3 December 2007 Abstract Hexokinases (HK) phosphorylate sugar immediately upon its entry into cells allowing these sugars to be metabolized. A total of four hexokinases have been characterized in a diversity of vertebrates—HKI, HKII, HKIII, and HKIV. HKIV is often called glucokinase (GCK) and has half the molecular weight of the other hexokinases, as it only has one hexokinase domain, while other vertebrate HKs have two. Differing hypothesis has been proposed to explain the diversification of the hexokinase gene family. We used a genomic approach to characterize hexokinase genes in a diverse array of vertebrate species and close relatives. Surprisingly we identified a fifth hexokinase-like gene, HKDC1 that exists and is expressed in diverse vertebrates. Analysis of the amino acid sequence of HKDC1 suggests that it may function as a hexokinase. To understand the evolution of the vertebrate hexokinase gene family we established a phylogeny of the hexokinase domain in all of the vertebrate hexokinase genes, as well as hexokinase genes from close relatives of the vertebrates. Our phylogeny demonstrates that duplication of the hexokinase domain, yielding a HK with two hexokinase domains, occurred prior to the diversification of the hexokinase gene family. -

Bioinformatics Analysis of HKDC1 Expression in Non-Small Cell Lung Cancer and Its Relationship to Survival
Journal of Nutritional Oncology, February 15, 2020, Volume 5, Number 1 DOI: 10.34175/jno202001004 · 31 · Bioinformatics Analysis of HKDC1 Expression in Non-small Cell Lung Cancer and Its Relationship to Survival Lei Wang, Wei Yuan Li, Fang Fang Li, Chun Yan Yang, Deng Cai Mu, Shang Yong Zheng School of Medicine, Yunnan University, Kunming, Yunnan 650091, China Abstract: Lung cancer has become one of the most common types of cancer, and has the highest morbidity and mortality rates. We herein performed a bioinformatic analysis to explore the gene expression of hexokinase domain containing 1 (HKDC1) to further illuminate the molecular mechanisms involved in non-small cell lung cancer (NSCLC). We also examined the relationship between HKDC1 expression and patient survival. The Oncomine and cBioPortal cancer genomics databases were used to investigate the expression of HKDC1 in NSCLC. Then the protein-protein interaction network and protein expression intensity of HKDC1 were examined with the Gene MANIA and The Human Protein Atlas databases. Finally, the prognostic impact of HKDC1 was evaluated via the GEPIA, Oncolnc and Kaplan-Meier Plotter online tools. HKDC1 was significantly up-regulated in NSCLC patients. In addition, the mutation frequency of HKDC1 in NSCLC was high in a large number of studies. Moreover, HKDC1 expression was correlated with a poor prognosis in NSCLC patients. These findings suggest that HKDC1 represents a novel therapeutic target for NSCLC. Key words: HKDC1; NSCLC; Bioinformatics; Expression; Carbohydrate metabolism Introduction genome-scale data source in life science research. Unlike Lung cancer (LC) is a life-threatening disease and the DNA sequence analysis and genotyping, microarrays most prominent cause of cancer morbidity and mortality in analyze mRNA. -

Mouse Models of Inherited Retinal Degeneration with Photoreceptor Cell Loss
cells Review Mouse Models of Inherited Retinal Degeneration with Photoreceptor Cell Loss 1, 1, 1 1,2,3 1 Gayle B. Collin y, Navdeep Gogna y, Bo Chang , Nattaya Damkham , Jai Pinkney , Lillian F. Hyde 1, Lisa Stone 1 , Jürgen K. Naggert 1 , Patsy M. Nishina 1,* and Mark P. Krebs 1,* 1 The Jackson Laboratory, Bar Harbor, Maine, ME 04609, USA; [email protected] (G.B.C.); [email protected] (N.G.); [email protected] (B.C.); [email protected] (N.D.); [email protected] (J.P.); [email protected] (L.F.H.); [email protected] (L.S.); [email protected] (J.K.N.) 2 Department of Immunology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok 10700, Thailand 3 Siriraj Center of Excellence for Stem Cell Research, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok 10700, Thailand * Correspondence: [email protected] (P.M.N.); [email protected] (M.P.K.); Tel.: +1-207-2886-383 (P.M.N.); +1-207-2886-000 (M.P.K.) These authors contributed equally to this work. y Received: 29 February 2020; Accepted: 7 April 2020; Published: 10 April 2020 Abstract: Inherited retinal degeneration (RD) leads to the impairment or loss of vision in millions of individuals worldwide, most frequently due to the loss of photoreceptor (PR) cells. Animal models, particularly the laboratory mouse, have been used to understand the pathogenic mechanisms that underlie PR cell loss and to explore therapies that may prevent, delay, or reverse RD. Here, we reviewed entries in the Mouse Genome Informatics and PubMed databases to compile a comprehensive list of monogenic mouse models in which PR cell loss is demonstrated. -

Gene Expression Profiling of Chromosome 10 in PTEN-Knockout
Gene Reports 21 (2020) 100895 Contents lists available at ScienceDirect Gene Reports journal homepage: www.elsevier.com/locate/genrep Gene expression profiling of chromosome 10 in PTEN-knockout (−/−) T human neural and mesenchymal stem cells: A system biology study ⁎ Hamid Fiujia, ,1, Mohammadreza Nassirib,1 a Department of Biochemistry, Faculty of Science, Payame Noor University of Mashhad (PNUM), Mashhad, Iran b Recombinant Protein Research Group, The Institute of Biotechnology, Ferdowsi University of Mashhad, Mashhad, Iran ARTICLE INFO ABSTRACT Keywords: The present study investigates the effects of PTEN deletion in human neuro and mesenchymal stem cells related PTEN deletion to the chromosome 10 gene expression profile. The RNA sequencing (RNA-seq) performed on four neural and NSCs four mesenchymal stem cells. DEG analysis outcome revealed 122 genes for neural stem cells (57 up-regulated MSCs and 65 down-regulated genes) and 258 genes for mesenchymal stem cells (98 up-regulated and 160 down- RNA sequencing regulated genes) that were deferentially expressed in the PTEN (-/-) group compared to the normal group. Gene Hub genes ontology analysis indicated that in the NSCs upregulated DEGs were significantly enriched in transcriptional Tumor progression activator activity, phosphatidylinositol phosphate phosphatase activity, MAP kinase activity while the down- regulated DEGs were mainly involved in glycolytic process through glucose-6-phosphate, canonical glycolysis, glucose catabolic process to pyruvate. Meanwhile, in MSCs, the upregulated -
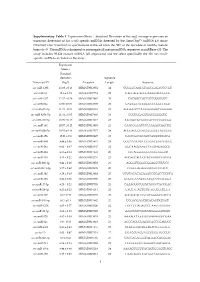
Supplementary Table 1. Expression
Supplementary Table 1. Expression (Mean Standard Deviation of the log2 average expression or transcript detection) of Sus scrofa specific miRNAs detected by the GeneChip™ miRNA 4.0 Array (ThermoFisher Scientific) in spermatozoa retrieved from the SRF of the ejaculate of healthy mature boars (n=3). The miRNA is designed to interrogate all mature miRNA sequences in miRBase v20. The array includes 30.424 mature miRNA (all organisms) and we select specifically the 326 Sus scrofa- specific miRNAs included in the array. Expression Mean ± Standard Deviation Sequence Transcript ID (log2) Accession Length Sequence ssc-miR-1285 13.98 ± 0.13 MIMAT0013954 24 CUGGGCAACAUAGCGAGACCCCGU ssc-miR-16 12.6 ± 0.74 MIMAT0007754 22 UAGCAGCACGUAAAUAUUGGCG ssc-miR-4332 12.32 ± 0.29 MIMAT0017962 20 CACGGCCGCCGCCGGGCGCC ssc-miR-92a 12.06 ± 0.09 MIMAT0013908 22 UAUUGCACUUGUCCCGGCCUGU ssc-miR-671-5p 11.73 ± 0.54 MIMAT0025381 24 AGGAAGCCCUGGAGGGGCUGGAGG ssc-miR-4334-5p 11.31 ± 0.05 MIMAT0017966 19 CCCUGGAGUGACGGGGGUG ssc-miR-425-5p 10.99 ± 0.15 MIMAT0013917 23 AAUGACACGAUCACUCCCGUUGA ssc-miR-191 10.57 ± 0.22 MIMAT0013876 23 CAACGGAAUCCCAAAAGCAGCUG ssc-miR-92b-5p 10.53 ± 0.18 MIMAT0017377 24 AGGGACGGGACGCGGUGCAGUGUU ssc-miR-15b 10.01 ± 0.9 MIMAT0002125 22 UAGCAGCACAUCAUGGUUUACA ssc-miR-30d 9.89 ± 0.36 MIMAT0013871 24 UGUAAACAUCCCCGACUGGAAGCU ssc-miR-26a 9.62 ± 0.47 MIMAT0002135 22 UUCAAGUAAUCCAGGAUAGGCU ssc-miR-484 9.55 ± 0.14 MIMAT0017974 20 CCCAGGGGGCGACCCAGGCU ssc-miR-103 9.53 ± 0.22 MIMAT0002154 23 AGCAGCAUUGUACAGGGCUAUGA ssc-miR-296-3p 9.41 ± 0.26 MIMAT0022958 -

New Insights Into Gestational Glucose Metabolism: Lessons Learned from 21St Century Approaches
Diabetes Volume 64, February 2015 327 Anthony R. Angueira,1 Anton E. Ludvik,1 Timothy E. Reddy,2,3 Barton Wicksteed,4 William L. Lowe Jr.,1 and Brian T. Layden1,5 New Insights Into Gestational Glucose Metabolism: Lessons Learned From 21st Century Approaches Diabetes 2015;64:327–334 | DOI: 10.2337/db14-0877 Pregnancy presents a unique physiological challenge that gestational diabetes mellitus (GDM) (i.e., new-onset hyper- requires changes coordinated by placentally and non– glycemia that presents during pregnancy) (1). Exemplifying PERSPECTIVES IN DIABETES placentally derived hormones to prepare the mother for the importance of studying GDM is that hyperglycemia the metabolic stress presented by fetal development and during pregnancy not only increases the risk of maternal to ensure appropriate nutrient allocation between mother type 2 diabetes (T2D), but also predisposes the developing and fetus. Of particular importance is the maintenance of fetus to poor metabolic health later in life (2). In this Per- normal glucose metabolism during pregnancy. Here, we spectives in Diabetes article, we first highlight key aspects of describe physiological changes in glucose metabolism normal gestational glucose metabolism. We then describe during pregnancy and highlight new insights into these new findings that have emerged in recent years spurred by adaptations that have emerged over the past decade us- new technologies (genome-wide association studies [GWAS], ing novel methodologies, specifically genome-wide asso- metabolomics, and gut microbiota investigations). Finally, ciation studies (GWAS) and metabolomics. While GWAS fi fi we place these ndings in context with current knowledge have identi ed some novel associations with metabolic fi traits during pregnancy, the majority of the findings over- in the eld and emphasize new directions emerging from lap with those observed in nonpregnant populations and these investigations. -

Genomic Analyses Reveal Recurrent Mutations in Epigenetic Modifiers
ARTICLE Received 6 Jul 2015 | Accepted 25 Aug 2015 | Published 29 Sep 2015 DOI: 10.1038/ncomms9470 OPEN Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK–STAT pathway in Se´zary syndrome Mark J. Kiel1,*, Anagh A. Sahasrabuddhe1,*,w, Delphine C.M. Rolland2,*, Thirunavukkarasu Velusamy1,*, Fuzon Chung1, Matthew Schaller1, Nathanael G. Bailey1, Bryan L. Betz1, Roberto N. Miranda3, Pierluigi Porcu4, John C. Byrd4, L. Jeffrey Medeiros3, Steven L. Kunkel1, David W. Bahler5, Megan S. Lim2 & Kojo S.J. Elenitoba-Johnson2,6 Se´zary syndrome (SS) is an aggressive leukaemia of mature T cells with poor prognosis and limited options for targeted therapies. The comprehensive genetic alterations underlying the pathogenesis of SS are unknown. Here we integrate whole-genome sequencing (n ¼ 6), whole-exome sequencing (n ¼ 66) and array comparative genomic hybridization-based copy- number analysis (n ¼ 80) of primary SS samples. We identify previously unknown recurrent loss-of-function aberrations targeting members of the chromatin remodelling/histone mod- ification and trithorax families, including ARID1A in which functional loss from nonsense and frameshift mutations and/or targeted deletions is observed in 40.3% of SS genomes. We also identify recurrent gain-of-function mutations targeting PLCG1 (9%) and JAK1, JAK3, STAT3 and STAT5B (JAK/STAT total B11%). Functional studies reveal sensitivity of JAK1-mutated primary SS cells to JAK inhibitor treatment. These results highlight the complex genomic landscape of SS and a role for inhibition of JAK/STAT pathways for the treatment of SS. 1 Department of Pathology, University of Michigan Medical School, Ann Arbor, Michigan 48109, USA. -

FUNCTIONAL CHARACTERIZATION of HEXOKINASE-LIKE 1 (HKL1) from ARABIDOPSIS THALIANA Abhijit Karve Clemson University, [email protected]
Clemson University TigerPrints All Dissertations Dissertations 12-2008 FUNCTIONAL CHARACTERIZATION OF HEXOKINASE-LIKE 1 (HKL1) FROM ARABIDOPSIS THALIANA Abhijit Karve Clemson University, [email protected] Follow this and additional works at: https://tigerprints.clemson.edu/all_dissertations Part of the Genetics Commons Recommended Citation Karve, Abhijit, "FUNCTIONAL CHARACTERIZATION OF HEXOKINASE-LIKE 1 (HKL1) FROM ARABIDOPSIS THALIANA" (2008). All Dissertations. 387. https://tigerprints.clemson.edu/all_dissertations/387 This Dissertation is brought to you for free and open access by the Dissertations at TigerPrints. It has been accepted for inclusion in All Dissertations by an authorized administrator of TigerPrints. For more information, please contact [email protected]. FUNCTIONAL CHARACTERIZATION OF HEXOKINASE-LIKE 1 (HKL1) FROM ARABIDOPSIS THALIANA A Thesis Presented to the Graduate School of Clemson University In Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy Genetics by Abhijit Avinash Karve December 2008 Accepted by: Dr. Brandon Moore, Committee Chair Dr. William Vance Baird Dr. Albert Abbott Dr. Lesly Temesvari Dr. James Morris ABSTRACT Arabidopsis hexokinase1 (AtHXK1) is a moonlighting protein with roles in both glucose signaling and catalysis. In this study, we first cloned and characterized the six HXK related genes from Arabidopsis. Three of the six encoded proteins were shown not to phosphorylate hexoses and thus, are designated as hexokinase-like (HKL) proteins. Though they are only 50% identical to HXK1, the amino acid sequences of HKL1 and HKL2 both have well conserved glucose binding domains and other recognized structural elements. The possible basis for their lack of catalytic activity was further probed by site-directed mutagenesis and ultimately was attributed to a suite of amino acid substitutions.