Centrin2 Regulates CP110 Removal in Primary Cilium Formation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information Proximity Interactions Among Centrosome
Current Biology, Volume 24 Supplemental Information Proximity Interactions among Centrosome Components Identify Regulators of Centriole Duplication Elif Nur Firat-Karalar, Navin Rauniyar, John R. Yates III, and Tim Stearns Figure S1 A Myc Streptavidin -tubulin Merge Myc Streptavidin -tubulin Merge BirA*-PLK4 BirA*-CEP63 BirA*- CEP192 BirA*- CEP152 - BirA*-CCDC67 BirA* CEP152 CPAP BirA*- B C Streptavidin PCM1 Merge Myc-BirA* -CEP63 PCM1 -tubulin Merge BirA*- CEP63 DMSO - BirA* CEP63 nocodazole BirA*- CCDC67 Figure S2 A GFP – + – + GFP-CEP152 + – + – Myc-CDK5RAP2 + + + + (225 kDa) Myc-CDK5RAP2 (216 kDa) GFP-CEP152 (27 kDa) GFP Input (5%) IP: GFP B GFP-CEP152 truncation proteins Inputs (5%) IP: GFP kDa 1-7481-10441-1290218-1654749-16541045-16541-7481-10441-1290218-1654749-16541045-1654 250- Myc-CDK5RAP2 150- 150- 100- 75- GFP-CEP152 Figure S3 A B CEP63 – – + – – + GFP CCDC14 KIAA0753 Centrosome + – – + – – GFP-CCDC14 CEP152 binding binding binding targeting – + – – + – GFP-KIAA0753 GFP-KIAA0753 (140 kDa) 1-496 N M C 150- 100- GFP-CCDC14 (115 kDa) 1-424 N M – 136-496 M C – 50- CEP63 (63 kDa) 1-135 N – 37- GFP (27 kDa) 136-424 M – kDa 425-496 C – – Inputs (2%) IP: GFP C GFP-CEP63 truncation proteins D GFP-CEP63 truncation proteins Inputs (5%) IP: GFP Inputs (5%) IP: GFP kDa kDa 1-135136-424425-4961-424136-496FL Ctl 1-135136-424425-4961-424136-496FL Ctl 1-135136-424425-4961-424136-496FL Ctl 1-135136-424425-4961-424136-496FL Ctl Myc- 150- Myc- 100- CCDC14 KIAA0753 100- 100- 75- 75- GFP- GFP- 50- CEP63 50- CEP63 37- 37- Figure S4 A siCtl -

The Transformation of the Centrosome Into the Basal Body: Similarities and Dissimilarities Between Somatic and Male Germ Cells and Their Relevance for Male Fertility
cells Review The Transformation of the Centrosome into the Basal Body: Similarities and Dissimilarities between Somatic and Male Germ Cells and Their Relevance for Male Fertility Constanza Tapia Contreras and Sigrid Hoyer-Fender * Göttingen Center of Molecular Biosciences, Johann-Friedrich-Blumenbach Institute for Zoology and Anthropology-Developmental Biology, Faculty of Biology and Psychology, Georg-August University of Göttingen, 37077 Göttingen, Germany; [email protected] * Correspondence: [email protected] Abstract: The sperm flagellum is essential for the transport of the genetic material toward the oocyte and thus the transmission of the genetic information to the next generation. During the haploid phase of spermatogenesis, i.e., spermiogenesis, a morphological and molecular restructuring of the male germ cell, the round spermatid, takes place that includes the silencing and compaction of the nucleus, the formation of the acrosomal vesicle from the Golgi apparatus, the formation of the sperm tail, and, finally, the shedding of excessive cytoplasm. Sperm tail formation starts in the round spermatid stage when the pair of centrioles moves toward the posterior pole of the nucleus. The sperm tail, eventually, becomes located opposed to the acrosomal vesicle, which develops at the anterior pole of the nucleus. The centriole pair tightly attaches to the nucleus, forming a nuclear membrane indentation. An Citation: Tapia Contreras, C.; articular structure is formed around the centriole pair known as the connecting piece, situated in the Hoyer-Fender, S. The Transformation neck region and linking the sperm head to the tail, also named the head-to-tail coupling apparatus or, of the Centrosome into the Basal in short, HTCA. -

Anchoring the Pole Plasm
RESEARCH HIGHLIGHTS Anchoring the pole plasm A kinesin in ciliogenesis Ectopic expression of Kif24 specifically desta- bilizes centriolar, but not cytoplasmic, microtu- The pole plasm, a cytoplasmic region contain- bules. In vitro experiments confirm the ability of ing maternal mRNAs and proteins at the pos- Kif24 to bind and depolymerize microtubules. terior of Drosophila oocytes, is essential for Furthermore, Kif24 overexpression suppresses germline and abdominal development. Pole cilia formation in starved cells, but, surprisingly, plasm assembly is initiated by the microtubule- also suppresses centriole elongation induced by dependent transport of oskar (osk) mRNA to Cep97 depletion. In both cases the microtubule the posterior, where the Osk protein stimulates depolymerizing domain of Kif24 is required. endocytic and actin-remodelling events essen- Thus, the authors suggest that Kif24 sup- tial for germ plasm functionality. Nakamura and presses cilia assembly from the mother cen- colleagues (Development 138, 2523–2532; 2011) triole through two mechanisms: by stabilizing have found that Mon2, a protein associated with or recruiting CP110 and through microtubule Golgi and endosomes, acts downstream of Osk remodelling. CKR to remodel cortical actin and anchor the pole In serum-starved cells, the primary cilium plasm. mon2 was identified in a screen for genes is nucleated from the mother centriole. required for the localization of the pole plasm Both cilia formation and centriole elonga- component Vasa. The authors found that Osk- tion is restricted by the centrosomal protein miRNA control of glucose induced formation of actin protrusions was CP110. Brian Dynlacht and co-workers have metabolism perturbed in mon2 mutants, as seen previously now identified the kinesin Kif24 as a CP110- in endosomal GTPase rab5 mutants. -

PPP1R35 Is a Novel Centrosomal Protein That Regulates
RESEARCH ARTICLE PPP1R35 is a novel centrosomal protein that regulates centriole length in concert with the microcephaly protein RTTN Andrew Michael Sydor1, Etienne Coyaud2, Cristina Rovelli1, Estelle Laurent2, Helen Liu1, Brian Raught2,3, Vito Mennella1,4* 1Cell Biology Program, The Hospital for Sick Children, Toronto, Canada; 2Princess Margaret Cancer Centre, University Health Network, Toronto, Canada; 3Department of Medical Biophysics, University of Toronto, Ontario, Canada; 4Department of Biochemistry, University of Toronto, Ontario, Canada Abstract Centrosome structure, function, and number are finely regulated at the cellular level to ensure normal mammalian development. Here, we characterize PPP1R35 as a novel bona fide centrosomal protein and demonstrate that it is critical for centriole elongation. Using quantitative super-resolution microscopy mapping and live-cell imaging we show that PPP1R35 is a resident centrosomal protein located in the proximal lumen above the cartwheel, a region of the centriole that has eluded detailed characterization. Loss of PPP1R35 function results in decreased centrosome number and shortened centrioles that lack centriolar distal and microtubule wall associated proteins required for centriole elongation. We further demonstrate that PPP1R35 acts downstream of, and forms a complex with, RTTN, a microcephaly protein required for distal centriole elongation. Altogether, our study identifies a novel step in the centriole elongation pathway centered on PPP1R35 and elucidates downstream partners of the microcephaly protein RTTN. DOI: https://doi.org/10.7554/eLife.37846.001 *For correspondence: [email protected] Introduction Competing interests: The The centrosome is a membrane-less organelle whose major role is to organize, orient, and regulate authors declare that no competing interests exist. the site of microtubule formation. -

Plk4-Induced Centriole Biogenesis in Human Cells
Plk4-induced Centriole Biogenesis in Human Cells Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften der Fakultät für Biologie der Ludwig-Maximilians Universität München Vorgelegt von Julia Kleylein-Sohn München, 2007 Dissertation eingereicht am: 27.11.2007 Tag der mündlichen Prüfung: 18.04.2008 Erstgutachter: Prof. E. A. Nigg Zweitgutachter: PD Dr. Angelika Böttger 2 Hiermit erkläre ich, dass ich die vorliegende Dissertation selbständig und ohne unerlaubte Hilfe angefertigt habe. Sämtliche Experimente wurden von mir selbst durchgeführt, soweit nicht explizit auf Dritte verwiesen wird. Ich habe weder an anderer Stelle versucht, eine Dissertation oder Teile einer solchen einzureichen bzw. einer Prüfungskommission vorzulegen, noch eine Doktorprüfung zu absolvieren. München, den 22.11.2007 3 TABLE OF CONTENTS SUMMARY…………………………………………………………………………..………. 6 INTRODUCTION……………………………………………………………………………. 7 Structure of the centrosome…………………………………………………………….. 7 The centrosome cycle…………………………………………………………………..10 Kinases involved in the regulation of centriole duplication………………………….12 Maintenance of centrosome numbers………………………………………………...13 Licensing of centriole duplication……………………………………………………... 15 ‘De novo ’ centriole assembly pathways in mammalian cells…………………..…...15 Templated centriole biogenesis in mammalian cells……………………………….. 18 The role of centrins and Sfi1p in centrosome duplication ……………………...…..19 Centriole biogenesis in C. elegans …………………………………………………… 21 Centriole biogenesis in human cells………………………………………………….. 23 Centrosome -

Induction of Therapeutic Tissue Tolerance Foxp3 Expression Is
Downloaded from http://www.jimmunol.org/ by guest on October 2, 2021 is online at: average * The Journal of Immunology , 13 of which you can access for free at: 2012; 189:3947-3956; Prepublished online 17 from submission to initial decision 4 weeks from acceptance to publication September 2012; doi: 10.4049/jimmunol.1200449 http://www.jimmunol.org/content/189/8/3947 Foxp3 Expression Is Required for the Induction of Therapeutic Tissue Tolerance Frederico S. Regateiro, Ye Chen, Adrian R. Kendal, Robert Hilbrands, Elizabeth Adams, Stephen P. Cobbold, Jianbo Ma, Kristian G. Andersen, Alexander G. Betz, Mindy Zhang, Shruti Madhiwalla, Bruce Roberts, Herman Waldmann, Kathleen F. Nolan and Duncan Howie J Immunol cites 35 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/09/17/jimmunol.120044 9.DC1 This article http://www.jimmunol.org/content/189/8/3947.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 2, 2021. -

Human CPAP and CP110 in Centriole Elongation and Ciliogenesis
Human CPAP and CP110 in Centriole Elongation and Ciliogenesis Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften der Fakultät für Biologie der Ludwig-Maximilians Universität München Vorgelegt von Thorsten I. Schmidt München, 2010 Dissertation eingereicht am: 11.05.2010 Tag der mündlichen Prüfung: 25.10.2010 Erstgutachter: Prof. Dr. Erich A. Nigg Zweitgutachter: Prof. Dr. Angelika Böttger Hiermit erkläre ich, dass ich die vorliegende Dissertation selbständig und ohne unerlaubte Hilfe angefertigt habe. Sämtliche Experimente wurden von mir selbst durchgeführt, soweit nicht explizit auf Dritte verwiesen wird. Ich habe weder an anderer Stelle versucht, eine Dissertation oder Teile einer solchen einzureichen bzw. einer Prüfungskommission vorzulegen, noch eine Doktorprüfung zu absolvieren. München, den 11.05.2010 TABLE OF CONTENTS TABLE OF CONTENTS 1. SUMMARY............................................................................................................................1 2. INTRODUCTION .................................................................................................................2 2.1 Function and Structure of the Centrosome.....................................................................2 2.1.1 The Centrosome as MTOC in Proliferating Cells .................................................2 2.1.2 The Centriole as Template for Cilia and Flagella .................................................3 2.1.3 Molecular Composition and Structure of the Centrosome....................................3 2.2 -

Ciliary Transcription Factors and Mirnas Precisely Regulate Cp110
RESEARCH ARTICLE Ciliary transcription factors and miRNAs precisely regulate Cp110 levels required for ciliary adhesions and ciliogenesis Peter Walentek1*, Ian K Quigley2, Dingyuan I Sun1, Umeet K Sajjan1, Christopher Kintner2, Richard M Harland1* 1Division of Genetics, Genomics and Development, Center for Integrative Genomics, Department of Molecular and Cell Biology, University of California, Berkeley, United States; 2Molecular Neurobiology Laboratory, Salk Institute for Biological Studies, La Jolla, United States Abstract Upon cell cycle exit, centriole-to-basal body transition facilitates cilia formation. The centriolar protein Cp110 is a regulator of this process and cilia inhibitor, but its positive roles in ciliogenesis remain poorly understood. Using Xenopus we show that Cp110 inhibits cilia formation at high levels, while optimal levels promote ciliogenesis. Cp110 localizes to cilia-forming basal bodies and rootlets, and is required for ciliary adhesion complexes that facilitate Actin interactions. The opposing roles of Cp110 in ciliation are generated in part by coiled-coil domains that mediate preferential binding to centrioles over rootlets. Because of its dual role in ciliogenesis, Cp110 levels must be precisely controlled. In multiciliated cells, this is achieved by both transcriptional and post- transcriptional regulation through ciliary transcription factors and microRNAs, which activate and repress cp110 to produce optimal Cp110 levels during ciliogenesis. Our data provide novel insights into how Cp110 and its regulation contribute to development and cell function. DOI: 10.7554/eLife.17557.001 *For correspondence: walentek@ berkeley.edu (PW); harland@ berkeley.edu (RMH) Introduction Competing interests: The Cilia are membrane-covered cell protrusions containing an axoneme of microtubules. Modified cen- authors declare that no trioles, called basal bodies, dock to the cell membrane, serve as microtubule organizing centers competing interests exist. -

Table SV. GO and KEGG Analysis of the Co-Expressed Pcgs with Predicting Pcgs and Lncrnas by Clusterprofiler
Table SV. GO and KEGG analysis of the co-expressed PCGs with predicting PCGs and lncRNAs by clusterProfiler. ONTOLOGY ID Description GeneRati P-value p adjust q value Count SYMBOL_ID o KEGG hsa03040 Spliceosome 22/336 < 0.001 0.00001 0.00001 22 BCAS2, DDX42, DDX46, DHX15, HNRNPK, LSM5, MAGOH, PLRG1, PPIE, PRPF18, PRPF38A, PRPF8, RBM8A, SF3A1, SF3B2, SNRNP200, SNRPD1, SNRPD3, SNRPE, SNRPF, SRSF1, SRSF6 CC GO:0098798 Mitochondrial 36/947 < 0.001 0.00002 0.00002 36 APOO, BCKDHB, GRPEL1, IMMP1L, MRPL27, MRPL30, MRPL35, MRPL49, MRPL50, MRPL57, protein complex MRPS14, MRPS21, MRPS31, MRPS33, MTERF4, NDUFA12, NDUFA13, NDUFA5, NDUFA6, NDUFA7, NDUFA8, NDUFB1, NDUFB6, NDUFC2, PARK7, PMPCB, SMDT1, SPG7, TIMM13, TIMM21, TIMM22, UQCC3, UQCRFS1, UQCRH, UQCRHL, COX7C CC GO:0031301 Integral 28/947 < 0.001 0.00002 0.00002 28 CHST12, LEMD2, TVP23C, APOO, ATP6V1G2, B4GAT1, CASD1, FUNDC2, ITM2B, L2HGDH, MFF, component of MPC2, PEX10, PEX11B, PEX16, SCO1, SLC22A17, SLC25A4, SLC35B1, SMDT1, SPG7, STEAP2, SV2A, organelle SYP, SYT4, TVP23B, UBIAD1, UQCC3 membrane CC GO:0005684 U2-type 15/947 < 0.001 0.00004 0.00004 15 BCAS2, CWC22, DHX15, GCFC2, LUC7L3, PLRG1, PPIE, PRPF18, PRPF8, SF3A1, SF3B2, SNRPD1, spliceosomal SNRPD3, SNRPE, SNRPF complex CC GO:0005681 Spliceosomal 28/947 < 0.001 0.00005 0.00005 28 BCAS2, CWC22, DDX25, DHX15, GCFC2, HNRNPH3, HNRNPK, HNRNPR, IK, LSM5, LUC7L3, complex MAGOH, PLRG1, PPIE, PRPF18, PRPF38A, PRPF8, RBM8A, SF3A1, SF3B2, SNRNP200, SNRPD1, SNRPD3, SNRPE, SNRPF, SRSF1, SYNCRIP, WBP4 CC GO:0031300 Intrinsic 28/947 < 0.001 0.00005 0.00005 -

Supplemental Solier
Supplementary Figure 1. Importance of Exon numbers for transcript downregulation by CPT Numbers of down-regulated genes for four groups of comparable size genes, differing only by the number of exons. Supplementary Figure 2. CPT up-regulates the p53 signaling pathway genes A, List of the GO categories for the up-regulated genes in CPT-treated HCT116 cells (p<0.05). In bold: GO category also present for the genes that are up-regulated in CPT- treated MCF7 cells. B, List of the up-regulated genes in both CPT-treated HCT116 cells and CPT-treated MCF7 cells (CPT 4 h). C, RT-PCR showing the effect of CPT on JUN and H2AFJ transcripts. Control cells were exposed to DMSO. β2 microglobulin (β2) mRNA was used as control. Supplementary Figure 3. Down-regulation of RNA degradation-related genes after CPT treatment A, “RNA degradation” pathway from KEGG. The genes with “red stars” were down- regulated genes after CPT treatment. B, Affy Exon array data for the “CNOT” genes. The log2 difference for the “CNOT” genes expression depending on CPT treatment was normalized to the untreated controls. C, RT-PCR showing the effect of CPT on “CNOT” genes down-regulation. HCT116 cells were treated with CPT (10 µM, 20 h) and CNOT6L, CNOT2, CNOT4 and CNOT6 mRNA were analysed by RT-PCR. Control cells were exposed to DMSO. β2 microglobulin (β2) mRNA was used as control. D, CNOT6L down-regulation after CPT treatment. CNOT6L transcript was analysed by Q- PCR. Supplementary Figure 4. Down-regulation of ubiquitin-related genes after CPT treatment A, “Ubiquitin-mediated proteolysis” pathway from KEGG. -

Detection of H3k4me3 Identifies Neurohiv Signatures, Genomic
viruses Article Detection of H3K4me3 Identifies NeuroHIV Signatures, Genomic Effects of Methamphetamine and Addiction Pathways in Postmortem HIV+ Brain Specimens that Are Not Amenable to Transcriptome Analysis Liana Basova 1, Alexander Lindsey 1, Anne Marie McGovern 1, Ronald J. Ellis 2 and Maria Cecilia Garibaldi Marcondes 1,* 1 San Diego Biomedical Research Institute, San Diego, CA 92121, USA; [email protected] (L.B.); [email protected] (A.L.); [email protected] (A.M.M.) 2 Departments of Neurosciences and Psychiatry, University of California San Diego, San Diego, CA 92103, USA; [email protected] * Correspondence: [email protected] Abstract: Human postmortem specimens are extremely valuable resources for investigating trans- lational hypotheses. Tissue repositories collect clinically assessed specimens from people with and without HIV, including age, viral load, treatments, substance use patterns and cognitive functions. One challenge is the limited number of specimens suitable for transcriptional studies, mainly due to poor RNA quality resulting from long postmortem intervals. We hypothesized that epigenomic Citation: Basova, L.; Lindsey, A.; signatures would be more stable than RNA for assessing global changes associated with outcomes McGovern, A.M.; Ellis, R.J.; of interest. We found that H3K27Ac or RNA Polymerase (Pol) were not consistently detected by Marcondes, M.C.G. Detection of H3K4me3 Identifies NeuroHIV Chromatin Immunoprecipitation (ChIP), while the enhancer H3K4me3 histone modification was Signatures, Genomic Effects of abundant and stable up to the 72 h postmortem. We tested our ability to use H3K4me3 in human Methamphetamine and Addiction prefrontal cortex from HIV+ individuals meeting criteria for methamphetamine use disorder or not Pathways in Postmortem HIV+ Brain (Meth +/−) which exhibited poor RNA quality and were not suitable for transcriptional profiling. -
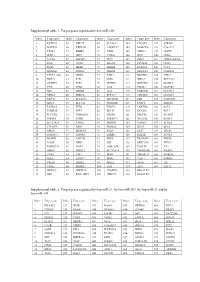
Target Genes Regulated by Hsa-Mir-21, by Hsa-Mir-203, by Hsa-Mir-21 and by Hsa-Mir-143
Supplemental table 1: Target genes regulated by hsa-miR-205 Index Target gene Index Target gene Index Target gene Index Target gene Index Target gene 1 KCTD20 35 UBE2Z 69 SLC38A1 103 LPCAT1 137 STK38L 2 MAPK14 36 YWHAH 70 ANGPTL7 104 MARCKS 138 C1orf123 3 TXNL1 37 RBBP4 71 CTGF 105 MED13 139 GUCD1 4 SPDL1 38 LRP1 72 CYR61 106 IPO7 140 CDK6 5 TCF20 39 IMPAD1 73 TP73 107 PHC2 141 CDKN2AIPNL 6 RAN 40 GNAS 74 EGLN2 108 PICALM 142 CLIP1 7 RGS6 41 MED1 75 ERBB2 109 PLAGL2 143 CUL5 8 HOXA11 42 INPPL1 76 PRRG4 110 NDUFA4 144 C6orf201 9 PAPPA-AS1 43 DDX5 77 F2RL2 111 NDUFB2 145 VTI1A 10 PRR15 44 E2F1 78 GOT1 112 NIPA2 146 SLC5A12 11 ACTRT3 45 E2F5 79 NUFIP2 113 NOTCH2 147 MAML2 12 YES1 46 ZEB2 80 IL24 114 PANK1 148 MAP3K9 13 SRC 47 ERBB3 81 IL32 115 PARD6B 149 NUDT21 14 NPRL3 48 PRKCE 82 RNF217 116 TMEM66 150 DNAJA1 15 NFAT5 49 SLC41A1 83 ZNF585B 117 EZR 151 CCDC108 16 XPOT 50 SLC7A2 84 SIGMAR1 118 ENPP4 152 SHISA6 17 KCTD16 51 ZEB1 85 VEGFA 119 LRRTM4 153 ACP1 18 TMSB4X 52 PHF8 86 BCL9L 120 KCNJ10 154 BCL2 19 PLCXD2 53 TMEM201 87 CREB1 121 PHLPP2 155 NCAPG 20 TNFSF8 54 PTPRJ 88 SERINC3 122 YEATS2 156 KLHL5 21 SLC25A25 55 ETNK1 89 HMGB3 123 VAMP1 157 ACSL4 22 C11orf74 56 XPR1 90 SRD5A1 124 RTN3 158 BCL6 23 GM2A 57 MRPL44 91 PTEN 125 RFX7 159 ITGA5 24 SMNDC1 58 TM9SF2 92 ESRRG 126 RAP2B 160 ACSL1 25 BAMBI 59 PAIP2B 93 PRLR 127 TRAF3IP1 161 EID2B 26 LCOR 60 NEK9 94 ICK 128 SERTAD2 162 TEX35 27 TMEM239 61 NOX5 95 LOH12CR1 129 TOLLIP 163 YY1 28 AMOT 62 DMXL2 96 SLC39A14 130 TMEM55B 164 SMAD1 29 CDK1 63 ETF1 97 BDP1 131 TMEM123 165 SMAD4 30 SQLE 64