NAP1L1 Is a Prognostic Biomarker and Contribute to Doxorubicin
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Macrophage Activation JUNB Is a Key Transcriptional Modulator Of
JUNB Is a Key Transcriptional Modulator of Macrophage Activation Mary F. Fontana, Alyssa Baccarella, Nidhi Pancholi, Miles A. Pufall, De'Broski R. Herbert and Charles C. Kim This information is current as of October 2, 2021. J Immunol 2015; 194:177-186; Prepublished online 3 December 2014; doi: 10.4049/jimmunol.1401595 http://www.jimmunol.org/content/194/1/177 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2014/12/03/jimmunol.140159 Material 5.DCSupplemental References This article cites 40 articles, 7 of which you can access for free at: http://www.jimmunol.org/content/194/1/177.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 2, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2014 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology JUNB Is a Key Transcriptional Modulator of Macrophage Activation Mary F. Fontana,* Alyssa Baccarella,* Nidhi Pancholi,* Miles A. -

NAP1L1: a Novel Human Colorectal Cancer Biomarker Derived from Animal Models of Apc Inactivation
fonc-10-01565 August 7, 2020 Time: 19:2 # 1 ORIGINAL RESEARCH published: 11 August 2020 doi: 10.3389/fonc.2020.01565 NAP1L1: A Novel Human Colorectal Cancer Biomarker Derived From Animal Models of Apc Inactivation Cleberson J. S. Queiroz1,2, Fei Song1,3, Karen R. Reed4,5, Nadeem Al-Khafaji1, Alan R. Clarke5†, Dale Vimalachandran6, Fabio Miyajima1,7, D. Mark Pritchard1* and John R. Jenkins1 1 Institute of Systems, Molecular and Integrative Biology, Henry Wellcome Laboratory, University of Liverpool, Liverpool, United Kingdom, 2 Faculty of Medicine, Federal University of Mato Grosso (UFMT), Cuiaba, Brazil, 3 INFRAFRONTIER GmbH, Neuherberg, Germany, 4 Wales Gene Park, Division of Cancer and Genetics, Cardiff University School of Medicine, Cardiff, United Kingdom, 5 European Cancer Stem Cell Research Institute, Cardiff University School of Biosciences, Cardiff, United Kingdom, 6 Department of Colorectal Surgery, Countess of Chester Hospital NHS Foundation Trust, Chester, Edited by: United Kingdom, 7 Molecular Epidemiology Laboratory, Oswaldo Cruz Foundation, Eusebio, Brazil Boris Zhivotovsky, Karolinska Institutet (KI), Sweden Introduction: Colorectal cancer (CRC) is the second leading cause of cancer death Reviewed by: Giovanna Caderni, worldwide and most deaths result from metastases. We have analyzed animal models University of Florence, Italy in which Apc, a gene that is frequently mutated during the early stages of colorectal Gabriele Multhoff, carcinogenesis, was inactivated and human samples to try to identify novel potential Technical University of Munich, Germany biomarkers for CRC. *Correspondence: Materials and Methods: We initially compared the proteomic and transcriptomic D. Mark Pritchard [email protected]; profiles of the small intestinal epithelium of transgenic mice in which Apc and/or Myc [email protected] had been inactivated. -

NAP1L1-MLLT10 Is a Rare Recurrent Translocation That Is Associated With
NAP1L1-MLLT10 is a rare recurrent translocation that is associated with HOXA activation and poor treatment response in T-cell acute lymphoblastic leukaemia Jonathan Bond, Aurore Touzart, Agata Cieslak, Amélie Trinquand, Tony Marchand, Martine Escoffre, Audrey Contet, Marc Muller, Claudine Schmitt, Thierry Fest, et al. To cite this version: Jonathan Bond, Aurore Touzart, Agata Cieslak, Amélie Trinquand, Tony Marchand, et al.. NAP1L1- MLLT10 is a rare recurrent translocation that is associated with HOXA activation and poor treatment response in T-cell acute lymphoblastic leukaemia. British Journal of Haematology, Wiley, 2016, 174 (3), pp.470-473. 10.1111/bjh.13772. hal-01217983 HAL Id: hal-01217983 https://hal-univ-rennes1.archives-ouvertes.fr/hal-01217983 Submitted on 14 Jan 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. NAP1L1-MLLT10 is a rare recurrent translocation that is associated with HOXA activation and poor treatment response in T-cell acute lymphoblastic leukaemia. Correspondence Jonathan Bond1,2, Aurore Touzart1,2, Agata Cieslak1,2, -

Nucleosome Eviction and Activated Transcription Require P300 Acetylation of Histone H3 Lysine 14
Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14 Whitney R. Luebben1, Neelam Sharma1, and Jennifer K. Nyborg2 Department of Biochemistry and Molecular Biology, Campus Box 1870, Colorado State University, Fort Collins, CO 80523 Edited by Mark T. Groudine, Fred Hutchinson Cancer Research Center, Seattle, WA, and approved September 9, 2010 (received for review July 6, 2010) Histone posttranslational modifications and chromatin dynamics include H4 acetylation-dependent relaxation of the chromatin are inextricably linked to eukaryotic gene expression. Among fiber and recruitment of ATP-dependent chromatin-remodeling the many modifications that have been characterized, histone tail complexes via acetyl-lysine binding bromodomains (12–17). acetylation is most strongly correlated with transcriptional activa- However, the mechanism by which histone tail acetylation elicits tion. In Metazoa, promoters of transcriptionally active genes are chromatin reconfiguration and coupled transcriptional activation generally devoid of physically repressive nucleosomes, consistent is unknown (6). with the contemporaneous binding of the large RNA polymerase II In recent years, numerous high profile mapping studies of transcription machinery. The histone acetyltransferase p300 is also nucleosomes and their modifications identified nucleosome-free detected at active gene promoters, flanked by regions of histone regions (NFRs) at the promoters of transcriptionally active genes hyperacetylation. Although the correlation -

Histone Variants — Ancient Wrap Artists of the Epigenome
REVIEWS CHROMATIN DYNAMICS Histone variants — ancient wrap artists of the epigenome Paul B. Talbert and Steven Henikoff Abstract | Histones wrap DNA to form nucleosome particles that compact eukaryotic genomes. Variant histones have evolved crucial roles in chromosome segregation, transcriptional regulation, DNA repair, sperm packaging and other processes. ‘Universal’ histone variants emerged early in eukaryotic evolution and were later displaced for bulk packaging roles by the canonical histones (H2A, H2B, H3 and H4), the synthesis of which is coupled to DNA replication. Further specializations of histone variants have evolved in some lineages to perform additional tasks. Differences among histone variants in their stability, DNA wrapping, specialized domains that regulate access to DNA, and post-translational modifications, underlie the diverse functions that histones have acquired in evolution. Histone chaperone Nearly all eukaryotes wrap their DNA around histones to In this Review, we consider the diversity and evolution of An escort protein that form nucleosomes that compact the genome while still core histone variants, from archaeal ancestors to univer- performs a transfer reaction on allowing access for active processes such as trans cription, sal eukaryotic variants to lineage-specific variants and a histone, such as deposition replication and DNA repair. Each nucleosome core variant-specific post-translational modifications (PTMs). onto DNA, eviction from DNA, transfer to another chaperone particle comprises ~ 147 bp of DNA wrapped in 1.7 turns We summarize their diverse and dynamic interactions as or enzyme, or storage for later around a protein octamer of 2 molecules of each of the 4 ‘wrap artists’ of DNA and discuss recent developments use. -

JAZF1, a Novel P400/TIP60/Nua4 Complex Member, Regulates H2A.Z Acetylation at Regulatory Regions
International Journal of Molecular Sciences Article JAZF1, A Novel p400/TIP60/NuA4 Complex Member, Regulates H2A.Z Acetylation at Regulatory Regions Tara Procida 1, Tobias Friedrich 1,2, Antonia P. M. Jack 3,†, Martina Peritore 3,‡ , Clemens Bönisch 3,§, H. Christian Eberl 4,k , Nadine Daus 1, Konstantin Kletenkov 1, Andrea Nist 5, Thorsten Stiewe 5 , Tilman Borggrefe 2, Matthias Mann 4, Marek Bartkuhn 1,6,* and Sandra B. Hake 1,3,* 1 Institute for Genetics, Justus-Liebig University Giessen, 35392 Giessen, Germany; [email protected] (T.P.); [email protected] (T.F.); [email protected] (N.D.); [email protected] (K.K.) 2 Institute for Biochemistry, Justus-Liebig-University Giessen, 35392 Giessen, Germany; [email protected] 3 Department of Molecular Biology, BioMedical Center (BMC), Ludwig-Maximilians-University Munich, 82152 Planegg-Martinsried, Germany; [email protected] (A.P.M.J.); [email protected] (M.P.); [email protected] (C.B.) 4 Department of Proteomics and Signal Transduction, Max Planck Institute of Biochemistry, 82152 Martinsried, Germany; [email protected] (H.C.E.); [email protected] (M.M.) 5 Genomics Core Facility, Institute of Molecular Oncology, Member of the German Center for Lung Research (DZL), Philipps-University Marburg, 35043 Marburg, Germany; [email protected] (A.N.); [email protected] (T.S.) 6 Biomedical Informatics and Systems Medicine, Justus-Liebig-University Giessen, 35392 Giessen, Germany * Correspondence: [email protected] (M.B.); [email protected] (S.B.H.); Tel.: +49-(0)641-99-30522 (M.B.); +49-(0)641-99-35460 (S.B.H.); Fax: +49-(0)641-99-35469 (S.B.H.) † Current Address: Sandoz/Hexal AG, 83607 Holzkirchen, Germany. -

DF7076-NAP1L1 Antibody
Affinity Biosciences website:www.affbiotech.com order:[email protected] NAP1L1 Antibody Cat.#: DF7076 Concn.: 1mg/ml Mol.Wt.: 45kDa,60kDa Size: 50ul,100ul,200ul Source: Rabbit Clonality: Polyclonal Application: WB 1:500-1:2000, IHC 1:50-1:200, ELISA(peptide) 1:20000-1:40000 *The optimal dilutions should be determined by the end user. Reactivity: Human,Mouse,Rat Purification: The antiserum was purified by peptide affinity chromatography using SulfoLink™ Coupling Resin (Thermo Fisher Scientific). Specificity: NAP1L1 Antibody detects endogenous levels of total NAP1L1. Immunogen: A synthesized peptide derived from human NAP1L1, corresponding to a region within C-terminal amino acids. Uniprot: P55209 Description: This gene encodes a member of the nucleosome assembly protein (NAP) family. This protein participates in DNA replication and may play a role in modulating chromatin formation and contribute to the regulation of cell proliferation. Alternative splicing of this gene results in several transcript variants; however, not all have been fully described. Storage Condition and Rabbit IgG in phosphate buffered saline , pH 7.4, 150mM Buffer: NaCl, 0.02% sodium azide and 50% glycerol.Store at -20 °C.Stable for 12 months from date of receipt. Western blot analysis of extracts from 293, using NAP1L1 Antibody. The lane on the left was treated with blocking peptide. Observed bands: 60 kDa. 1 / 2 Affinity Biosciences website:www.affbiotech.com order:[email protected] Western blot analysis of extracts from mouse thymus tissue lysates, using NAP1L1 antibody. The lane on the left was treated with the antigen-specific peptide. DF7076 at 1/100 staining Rat liver tissue by IHC-P. -

Differential Action on Transcriptional Programs Related to Cell Cycle Control and Immune Function
Oncogene (2007) 26, 6307–6318 & 2007 Nature Publishing Group All rights reserved 0950-9232/07 $30.00 www.nature.com/onc ORIGINAL ARTICLE Loss of the retinoblastoma tumor suppressor: differential action on transcriptional programs related to cell cycle control and immune function MP Markey1, J Bergseid2, EE Bosco1, K Stengel1,HXu3,4, CN Mayhew1, SJ Schwemberger6, WA Braden1, Y Jiang2, GF Babcock5,6, AG Jegga3,4, BJ Aronow3,4, MF Reed5, JYJ Wang2 and ES Knudsen1 1Department of Cell and Cancer Biology, University of Cincinnati, Cincinnati, OH, USA; 2Department of Medicine, Moores Cancer Center, University of California San Diego, La Jolla, CA, USA; 3Department of Pediatrics, University of Cincinnati, Cincinnati, OH, USA; 4Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; 5Department of Surgery, University of Cincinnati, Cincinnati, OH, USA and 6Shriners Hospital, Cincinnati, OH, USA Functional inactivation of the retinoblastoma tumor genes involved in immune function was readily observed suppressor gene product (RB) is a common event in with disruption of the LXCXE-binding function of RB. human cancers. Classically, RB functions to constrain Thus, these studies demonstrate that RB plays a cellular proliferation, and loss of RB is proposed to significant role in both the positive and negative regula- facilitate the hyperplastic proliferation associated with tions of transcriptional programs and indicate that loss of tumorigenesis. To understand the repertoire of regulatory RB has distinct biological effects related to both cell cycle processes governed by RB, two models of RB loss were control and immune function. utilized to perform microarray analysis. In murine Oncogene (2007) 26, 6307–6318; doi:10.1038/sj.onc.1210450; embryonic fibroblasts harboring germline loss of RB, published online 23 April 2007 there was a striking deregulation of gene expression, wherein distinct biological pathways were altered. -

Pan-Cancer Multi-Omics Analysis and Orthogonal Experimental Assessment of Epigenetic Driver Genes
Downloaded from genome.cshlp.org on October 9, 2021 - Published by Cold Spring Harbor Laboratory Press Resource Pan-cancer multi-omics analysis and orthogonal experimental assessment of epigenetic driver genes Andrea Halaburkova, Vincent Cahais, Alexei Novoloaca, Mariana Gomes da Silva Araujo, Rita Khoueiry,1 Akram Ghantous,1 and Zdenko Herceg1 Epigenetics Group, International Agency for Research on Cancer (IARC), 69008 Lyon, France The recent identification of recurrently mutated epigenetic regulator genes (ERGs) supports their critical role in tumorigen- esis. We conducted a pan-cancer analysis integrating (epi)genome, transcriptome, and DNA methylome alterations in a cu- rated list of 426 ERGs across 33 cancer types, comprising 10,845 tumor and 730 normal tissues. We found that, in addition to mutations, copy number alterations in ERGs were more frequent than previously anticipated and tightly linked to ex- pression aberrations. Novel bioinformatics approaches, integrating the strengths of various driver prediction and multi- omics algorithms, and an orthogonal in vitro screen (CRISPR-Cas9) targeting all ERGs revealed genes with driver roles with- in and across malignancies and shared driver mechanisms operating across multiple cancer types and hallmarks. This is the largest and most comprehensive analysis thus far; it is also the first experimental effort to specifically identify ERG drivers (epidrivers) and characterize their deregulation and functional impact in oncogenic processes. [Supplemental material is available for this article.] Although it has long been known that human cancers harbor both gression, potentially acting as oncogenes or tumor suppressors genetic and epigenetic changes, with an intricate interplay be- (Plass et al. 2013; Vogelstein et al. 2013). -

RNA Binding Density on X-Chromosome Differing from That on 22 Autosomes in Human
RNA Binding Density on X-chromosome Differing from that on 22 Autosomes in Human LV Zhanjun*, LU Ying**, SONG Shuxia*, ZHAI Yu*,WANG Xiufang * *Department of Laboratory Animal, Hebei Medical University, Shijiazhuang 050017, China **Center for Theoretic Biology Peking University, Peking 100871, China Abstract To test whether X-chromosome has unique genomic characteristics, X-chromosome and 22 autosomes were compared for RNA binding density. Nucleotide sequences on the chromosomes were divided into 50kb per segment that was recoded as a set of frequency values of 7-nucleotide (7nt) strings using all possible 7nt strings (47=16384). 120 genes highly expressed in tonsil germinal center B cells were selected for calculating 7nt string frequency values of all introns (RNAs). The binding density of DNA segments and RNAs was determined by the amount of complement sequences. It was shown for the first time that gene-poor and low gene expression X-chromosome had the lowest percentage of the DNA segments that can highly bind RNAs, whereas gene-rich and high gene expression chromosome 19 had the highest percentage of the segments. On the basis of these results, it is proposed that the nonrandom properties of distribution of RNA highly binding DNA segments on the chromosomes provide strong evidence that lack of RNA highly binding segments may be a cause of X-chromosome inactivation. Key works: X-chromosome inactivation; intron RNA; DNA sequence composition; nucleotide string; Introduction Among placental mammals, the basic features of X-chromosome inactivation (XCI) are well established (BAILEY et al. 2000, OKAMOTO et al. 2000). XCI is the distinct difference between X-chromosome and autosomes. -
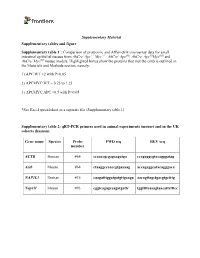
Supplementary Material Supplementary Tables and Figure Supplementary Table 1*: Comparison of Proteomic and Affymetrix Microarra
Supplementary Material Supplementary tables and figure Supplementary table 1*: Comparison of proteomic and Affymetrix microarray data for small intestinal epithelial tissues from AhCre+Apc+/+Myc+/+, AhCre+Apcfl/fl, AhCre+Apcfl/fMycfl/fll and AhCre+Mycfl/fl mouse models. Highlighted boxes show the proteins that met the criteria outlined in the Materials and Methods section, namely: 1) APC:WT >2 with P<0.05 2) APCMYC:WT = 0.75 to 1.25 3) APCMYC:APC <0.5 with P<0.05 *See Excel spreadsheet as a separate file (Supplementary table 1). Supplementary table 2: qRT-PCR primers used in animal experiments (mouse) and in the UK cohorts (human). Gene name Species Probe FWD seq REV seq number ACTB Human #64 ccaaccgcgagaagatga ccagaggcgtacagggatag Actb Mouse #64 ctaaggccaaccgtgaaaag accagaggcatacagggaca NAP1L1 Human #35 caagatttggatgatgttgaaga aacagttagctgacgtgctttg Nap1l1 Mouse #93 cggtcagagccagatgattc tggttttcaaagtaacattctttcc Supplementary Material Supplementary table 3: qRT-PCR assays used in the Brazil cohort. Gene • Entrez Gene Description • TaqMan® assay Id(*) Symbol ID 4673 nucleosome Hs00748775_s1 NAP1L1 assembly protein 1- like 1 60 actin, beta Hs99999903_m1 ACTB Supplementary figure 1. Comparison of NAP1L1 staining patterns observed with the IHC protocols used in the initial validation study (A) and in the prognostic study (B), using the same sample. Similar staining intensity and localisation are observed. Differences in colour shade and background are due to the use of different cameras for recording the images. Magnification: 600x. 2 Supplementary figure 2. IHC expression of NAP1L1 assessed manually using a modified H-score. This assessment was performed in a different cohort of patients, using a different scoring method. Samples were obtained from the Countess of Chester Hospital NHS Foundation Trust, Chester, UK.