Understanding PTEN Regulation: PIP2, Polarity and Protein Stability
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phosphatidylserine
Cognitive Vitality Reports® are reports written by neuroscientists at the Alzheimer’s Drug Discovery Foundation (ADDF). These scientific reports include analysis of drugs, drugs-in- development, drug targets, supplements, nutraceuticals, food/drink, non-pharmacologic interventions, and risk factors. Neuroscientists evaluate the potential benefit (or harm) for brain health, as well as for age-related health concerns that can affect brain health (e.g., cardiovascular diseases, cancers, diabetes/metabolic syndrome). In addition, these reports include evaluation of safety data, from clinical trials if available, and from preclinical models. Phosphatidylserine Evidence Summary May promote cognitive function and protect from decline, especially for DHA-enriched phosphatidylserine. It has a good safety profile but has limited bioavailability. Neuroprotective Benefit: Mixed evidence from clinical trials, and considerable bias in results reporting. Has poor bioavailability and it’s unclear how well it gets into the brain. Aging and related health concerns: No clear rationale or data. One study reported a minor increase in mobility in elderly, but effect can’t be clearly tied to phosphatidylserine. Safety: Well-tolerated with no serious adverse events reported in short trials. May slightly reduce blood pressure. Information on long-term safety is not available. 1 What are they? Phosphatidylserine (PS) is a class of phospholipids that help to make up the plasma membranes in the brain. Varying the levels and the symmetry of PS in cell membranes (i.e. on the inside or outside of a membrane) can affect signaling pathways that are central for cell survival (e.g. Akt, protein kinase C, and Raf-1) and neuronal synaptic communication [1]. -

Deciphering the Functions of Ets2, Pten and P53 in Stromal Fibroblasts in Multiple
Deciphering the Functions of Ets2, Pten and p53 in Stromal Fibroblasts in Multiple Breast Cancer Models DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Julie Wallace Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2013 Dissertation Committee: Michael C. Ostrowski, PhD, Advisor Gustavo Leone, PhD Denis Guttridge, PhD Dawn Chandler, PhD Copyright by Julie Wallace 2013 Abstract Breast cancer is the second most common cancer in American women, and is also the second leading cause of cancer death in women. It is estimated that nearly a quarter of a million new cases of invasive breast cancer will be diagnosed in women in the United States this year, and approximately 40,000 of these women will die from breast cancer. Although death rates have been on the decline for the past decade, there is still much we need to learn about this disease to improve prevention, detection and treatment strategies. The majority of early studies have focused on the malignant tumor cells themselves, and much has been learned concerning mutations, amplifications and other genetic and epigenetic alterations of these cells. However more recent work has acknowledged the strong influence of tumor stroma on the initiation, progression and recurrence of cancer. Under normal conditions this stroma has been shown to have protective effects against tumorigenesis, however the transformation of tumor cells manipulates this surrounding environment to actually promote malignancy. Fibroblasts in particular make up a significant portion of this stroma, and have been shown to impact various aspects of tumor cell biology. -

Biogenesis and Maintenance of Cytoplasmic Domains in Myelin of the Central Nervous System
Biogenesis and Maintenance of Cytoplasmic Domains in Myelin of the Central Nervous System Dissertation for the award of the degree “Doctor rerum naturalium” of the Georg-August-Universität Göttingen within the doctoral program Molecular Biology of Cells of the Georg-August University School of Science (GAUSS) submitted by Caroline Julia Velte from Usingen, Germany Göttingen 2016 Members of the Thesis Committee: Prof. Dr. Mikael Simons, Reviewer Max Planck Institute of Experimental Medicine, Göttingen Prof. Dr. Andreas Janshoff, Reviewer Georg-August University, Göttingen Prof. Dr. Dirk Görlich Max Planck Institute of Biophysical Chemistry, Göttingen Date of the thesis defense: 27th of June 2016 Don't part with your illusions. When they are gone, you may still exist, but you have ceased to live. (Mark Twain) III Affidavit I hereby declare that my PhD thesis “Biogenesis and Maintenance of Cytoplasmic Domains in Myelin of the Central Nervous System” has been written independently with no other aids or sources than quoted. Furthermore, I confirm that this thesis has not been submitted as part of another examination process neither in identical nor in similar form. Caroline Julia Velte April 2016 Göttingen, Germany V Publications Nicolas Snaidero*, Caroline Velte*, Matti Myllykoski, Arne Raasakka, Alexander Ignatev, Hauke B. Werner, Michelle S. Erwig, Wiebke Moebius, Petri Kursula, Klaus-Armin Nave, and Mikael Simons Antagonistic Functions of MBP and CNP Establish Cytosolic Channels in CNS Myelin, Cell Reports 18 *equal contribution (January 2017) E. d’Este, D. Kamin, C. Velte, F. Göttfert, M. Simons, S.W. Hell Subcortical cytoskeleton periodicity throughout the nervous system, Scientific Reports 6 (March 2016) K.A. -

Enzyme Phosphatidylserine Synthase (Saccharomyces Cerevisae/Chol Gene/Transformation) V
Proc. Nati. Acad. Sci. USA Vol. 80, pp. 7279-7283, December 1983 Genetics Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase (Saccharomyces cerevisae/CHOl gene/transformation) V. A. LETTS*, L. S. KLIG*, M. BAE-LEEt, G. M. CARMANt, AND S. A. HENRY* *Departments of Genetics and Molecular Biology, Albert Einstein College of Medicine, Bronx, NY 10461; and tDepartment of Food Science, Cook College, New Jersey Agricultural Experimental Station, Rutgers University, New Brunswick, NJ 08903 Communicated by Frank Lilly, August 11, 1983 ABSTRACT The structural gene (CHOI) for phosphatidyl- Mammals, for example, synthesize PtdSer by an exchange re- serine synthase (CDPdiacylglycerol:L-serine O-phosphatidyl- action with PtdEtn (9). However, PtdSer synthase is found in transferase, EC 2.7.8.8) was isolated by genetic complementation E. coli and indeed the structural gene for the E. coli enzyme has in Saccharomyces cerevmae from a bank of yeast genomic DNA been cloned (10). Thus, cloning of the structural gene for the on a chimeric plasmid. The cloned DNA (4.0 kilobases long) was yeast enzyme will permit a detailed comparison of the structure shown to represent a unique sequence in the yeast genome. The and function of prokaryotic and eukaryotic genes and gene DNA sequence on an integrative plasmid was shown to recombine products. The availability of a clone of the CHOI gene will per- into the CHOi locus, confwrming its genetic identity. The chol yeast mit analysis of its regulation at the transcriptional level. Fur- strain transformed with this gene on an autonomously replicating thermore, the cloning of the CHOI gene provides us with the plasmid had significantly increased activity of the regulated mem- the levels of PtdSer synthase in the cell, brane-associated enzyme phosphatidylserine synthase. -

Exploring the Plasmatic Platelet-Activating Factor Acetylhydrolase Activity in Patients with Anti-Phospholipid Antibodies
Autoimmun Highlights (2017) 8:5 https://doi.org/10.1007/s13317-017-0092-7 ORIGINAL ARTICLE Exploring the plasmatic platelet-activating factor acetylhydrolase activity in patients with anti-phospholipid antibodies 1,2 1 1 2 Martina Fabris • Adriana Cifu` • Cinzia Pistis • Massimo Siega-Ducaton • 1 1,2 3 1,2 Desre` Ethel Fontana • Roberta Giacomello • Elio Tonutti • Francesco Curcio Received: 23 December 2016 / Accepted: 12 March 2017 / Published online: 25 March 2017 Ó The Author(s) 2017. This article is an open access publication Abstract Conclusions PAF-AH plasmatic activity is particularly up- Purpose To explore the role of plasmatic platelet-activat- regulated in LAC? and in ab2GPI IgG? patients, possibly ing factor acetylhydrolase (PAF-AH), a marker of cardio- representing an alternative prognostic biomarker for the vascular risk, in patients with anti-phospholipid antibodies therapeutic management of APS patients. (aPL). Methods PAF-AH activity was assessed in a series of 167 Keywords Platelet-activating factor acetylhydrolase Á unselected patients screened for aPL in a context of Anti-phospholipid syndrome Á Anti-beta2-glycoprotein I thrombotic events, risk of thrombosis or obstetric compli- antibodies Á Anti-prothrombin/phosphatidylserine cations and in 77 blood donors. antibodies Á Lupus anticoagulant Á Atherosclerosis Results 116/167 patients showed positive results for at least one aPL among IgG/IgM anti-prothrombin/phos- phatidylserine (aPS/PT), anti-cardiolipin (aCL), anti-beta2- Introduction glycoprotein I (ab2GPI) or lupus anticoagulant (LAC), while 51/167 patients resulted aPL-negative. LAC? Anti-phospholipid syndrome (APS) is a hypercoagulable patients disclosed higher PAF-AH than LAC-negative disorder clinically displayed by venous or arterial throm- (22.1 ± 6.4 nmol/min/ml vs. -

Hypomethylation-Linked Activation of PLCE1 Impedes Autophagy and Promotes Tumorigenesis Through MDM2-Mediated Ubiquitination and Destabilization of P53
Author Manuscript Published OnlineFirst on February 17, 2020; DOI: 10.1158/0008-5472.CAN-19-1912 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Hypomethylation-linked activation of PLCE1 impedes autophagy and promotes tumorigenesis through MDM2-mediated ubiquitination and destabilization of p53 Yunzhao Chen1,3*, Huahua Xin1*, Hao Peng1*, Qi Shi1, Menglu Li1, Jie Yu3, Yanxia Tian1, Xueping Han1, Xi Chen1, Yi Zheng4, Jun Li5, Zhihao Yang1, Lan Yang1, Jianming Hu1, Xuan Huang2, Zheng Liu2, Xiaoxi Huang2, Hong Zhou6, Xiaobin Cui1**, Feng Li1,2** 1 Department of Pathology and Key Laboratory for Xinjiang Endemic and Ethnic Diseases, The First Affiliated Hospital, Shihezi University School of Medicine, Shihezi 832002, China; 2 Department of Pathology and Medical Research Center, Beijing Chaoyang Hospital, Capital Medical University, Beijing 100020, China; 3 The people's hospital of Suzhou National Hi-Tech District, Suzhou 215010, China; 4 Department of Gastroenterology, The First Affiliated Hospital, Shihezi University School of Medicine, Shihezi 832002, China; 5 Department of Ultrasound, The First Affiliated Hospital, Shihezi University School of Medicine, Shihezi 832002, China; 6 Bone Research Program, ANZAC Research Institute, The University of Sydney, Sydney, Australia. Running title: PLCE1 impedes autophagy via MDM2-p53 axis in ESCC *These authors contributed equally to this work. Corresponding authors: **Xiaobin Cui, Department of Pathology and Key Laboratory for Xinjiang Endemic and Ethnic Diseases, The First Affiliated Hospital, Shihezi University School of Medicine, Shihezi 832002, China. Phone: 86.0377.2850955; E-mail: [email protected]; **Feng Li, Department of Pathology and Medical Research Center, Beijing Chaoyang Hospital, Capital Medical University, Beijing 100020, China. -

Disruption of Epithelial Architecture Caused by Loss of PTEN Or by Oncogenic Mutant P110a/PIK3CA but Not by HER2 Or Mutant AKT1
Oncogene (2013) 32, 4417–4426 & 2013 Macmillan Publishers Limited All rights reserved 0950-9232/13 www.nature.com/onc ORIGINAL ARTICLE Disruption of epithelial architecture caused by loss of PTEN or by oncogenic mutant p110a/PIK3CA but not by HER2 or mutant AKT1 FM Berglund1,3, NR Weerasinghe1,3, L Davidson1,3, JC Lim1,{, BJ Eickholt2 and NR Leslie1 Genetic changes in HER2, PTEN, PIK3CA and AKT1 are all common in breast cancer and lead to the elevated phosphorylation of downstream targets of the PI3K/AKT signalling pathway. Changes in HER2, PTEN, PIK3CA and AKT have all been reported to lead to both enhanced proliferation and failures in hollow lumen formation in three dimensional epithelial culture models, but it is not clear whether these failures in lumen formation are caused by any failure in the spatial coordination of lumen formation (hollowing) or purely a failure in the apoptosis and clearance of luminal cells (cavitation). Here, we use normal murine mammary gland (NMuMG) epithelial cells, which form a hollow lumen without significant apoptosis, to compare the transformation by these four genetic changes. We find that either mutant PIK3CA expression or PTEN loss, but not mutant AKT1 E17K, cause disrupted epithelial architecture, whereas HER2 overexpression drives strong proliferation without affecting lumen formation in these cells. We also show that PTEN requires both lipid and protein phosphatase activity, its extreme C-terminal PDZ binding sequence and probably Myosin 5A to control lumen formation through a mechanism that does not correlate with its ability to control AKT, but which is selectively lost through mutation in some tumours. -

A Role for Nuclear PTEN in Neuronal Differentiation
The Journal of Neuroscience, February 15, 2000, 20(4):1404–1413 A Role for Nuclear PTEN in Neuronal Differentiation Mahesh B. Lachyankar,1 Nazneen Sultana,1 Christopher M. Schonhoff,2 Prasenjit Mitra,1 Wojciech Poluha,1 Stephen Lambert,3 Peter J. Quesenberry,4 N. Scott Litofsky,5 Lawrence D. Recht,6 Roya Nabi,7 Susan J. Miller,8 Shinji Ohta,8 Benjamin G. Neel,8 and Alonzo H. Ross1 Departments of 1Pharmacology and Molecular Toxicology, 2Biochemistry and Molecular Biology, 3Cell Biology, 4Medicine (Cancer Center), 5Surgery, and 6Neurology, University of Massachusetts Medical School, Worcester, Massachusetts 01655, 7Department of Biology and Biotechnology, Worcester Polytechnic Institute, Worcester, Massachusetts 01609, and 8Cancer Biology Program, Division of Hematology-Oncology, Department of Medicine, Beth Israel-Deaconess Medical Center, Boston, Massachusetts 02215 Mutations of phosphatase and tensin homolog deleted on chro- detected in both the nucleus and cytoplasm. Suppression of mosome 10 (PTEN), a protein and lipid phosphatase, have been PTEN levels with antisense oligonucleotides does not block associated with gliomas, macrocephaly, and mental deficien- initiation of neuronal differentiation. Instead, PTEN antisense cies. We have assessed PTENЈs role in the nervous system and leads to death of the resulting, immature neurons, probably find that PTEN is expressed in mouse brain late in develop- during neurite extension. In contrast, PTEN is not required for ment, starting at approximately postnatal day 0. In adult brain, astrocytic differentiation. These observations indicate that PTEN is preferentially expressed in neurons and is especially PTEN acts at multiple sites in the cell, regulating the transition evident in Purkinje neurons, olfactory mitral neurons, and large of differentiating neuroblasts to postmitotic neurons. -

Truseq 48 Gene Cancer Panel
TruSeq 48 Gene Cancer Panel Indication: Next generation sequencing (NGS) assay for detection of hot spot mutations in 48 cancer-related genes: ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, JAK2, JAK3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53 and VHL. The specific mutations are detected by amplification of the corresponding exons by polymerase chain reaction (PCR). The PCR product is sequenced on a Illumina's MiSeqDx NGS Sequencer. Annotation is provided by Cartagenia Bench Lab NGS. Clinical Panel Gene Exon / Amino Acid (AA) Coverage ABL1 Exons 4-7 (AA 243-362, 395-424) ALK Exons 23, 25 (AA 1172-1176, 1248-1275) BRAF Exon 15 (AA 581-606) EGFR Exons 3, 7, 18-21 (AA 108-142, 287-297, 598-627, 708-728, 729-761, 762-817, 857-875) GNA11 Exons 4-7 (AA 159-172, 172-202, 202-216, 255-297, 304-349, 349-360) GNAQ Exons 4- 7 (AA 159-202, 202-210, 210-263, 261-297, 297-325, 326-360, 355-360) IDH1 Exon 4 (AA 95-133) JAK2 Exon 14 (AA 615-622) KIT Exons 11, 13, 17 (AA 550-587, 641-664, 814-828) KRAS Exons 2-3 (AA 1-22, 38-63) MPL Exon 10 (AA 514-522) NRAS Exons 2-3 (AA 1-19, 38-62) NPM1 Exon 11 (AA 283-295) PIK3CA Exons 2, 5, 8, 10, 21 (AA 84-118, 345-353, 418-441, 538-555, 987-1068) Investigational Panel Gene Exon / Amino Acid (AA) Coverage AKT1 Exon 4 (AA 16-49) APC Exon 16 (AA 875-918, 1113-1153, 1257-1575) ATM Exons 8, 9, 12, 17, 26, 34-36, 50, 54-56, 59, -
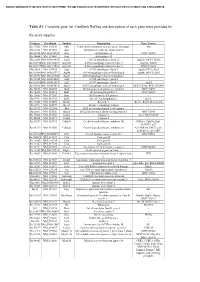
Table S1. Complete Gene List. Genbank Refseq and Description of Each Gene Were Provided By
Document downloaded from http://www.elsevier.es, day 24/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Table S1. Complete gene list. GenBank RefSeq and description of each gene were provided by the array supplier. Unigene GeneBank Symbol Description Gene Name/s Rn.11422 NM_033230 Akt1 V-akt murine thymoma viral oncogene homolog 1 Akt Rn.2104 NM_019288 App Amyloid beta (A4) precursor protein - Rn.23323 NM_001034933 Arsa Arylsulfatase A MGC125207 Rn.94004 NM_033443 Arsb Arylsulfatase B - Rn.6224 NM_001038495 Atg12 ATG12 autophagy related 12 Apg12l, MGC125080 Rn.101734NM_001108809 Atg16l1 ATG16 autophagy related 16-like 1 Apg16l, Wdr30 Rn.104199NM_001191560 Atg16l2 ATG16 autophagy related 16-like 2 RGD1311400 Rn.3084 NM_134394 Atg3 ATG3 autophagy related 3 Apg3l, PIG-1, Pig1 Rn.163086NM_001025711 Atg4b ATG4 autophagy related 4 homolog B Apg4b, MGC112887 Rn.23378 NM_001107948 Atg4c ATG4 autophagy related 4 homolog C - Rn.98385 NM_001014250 Atg5 ATG5 autophagy related 5 - Rn.162765NM_001012097 Atg7 ATG7 autophagy related 7 Apg7l Rn.35248 NM_001014218 Atg9a ATG9 autophagy related 9 homolog A MGC105908, RGD1310450 Rn.36696 NM_022698 Bad BCL2-associated agonist of cell death MGC72439 Rn.14598 NM_053812 Bak1 BCL2-antagonist/killer 1 MGC108627 Rn.10668 NM_017059 Bax Bcl2-associated X protein - Rn.9996 NM_016993 Bcl2 B-cell CLL/lymphoma 2 Bcl-2 Rn.10323 NM_031535 Bcl2l1 Bcl2-like 1 Bcl-xl, Bcl2l, Bclx, bcl-X Rn.2776 NM_053739 Becn1 Beclin 1, autophagy related - Rn.31142 NM_022684 -

The Functions of Tumor Suppressor PTEN in Innate and Adaptive Immunity
Cellular & Molecular Immunology (2017) 14, 581–589 & 2017 CSI and USTC All rights reserved 2042-0226/17 $32.00 www.nature.com/cmi REVIEW The functions of tumor suppressor PTEN in innate and adaptive immunity Lang Chen1 and Deyin Guo2 The tumor suppressor phosphatase and tensin homolog (PTEN) is a lipid and protein phosphatase that is able to antagonize the PI3K/AKT pathway and inhibit tumor growth. PTEN also possesses phosphatase-independent functions. Genetic alterations of PTEN may lead to the deregulation of cell proliferation, survival, differentiation, energy metabolism and cellular architecture and mobility. Although the role of PTEN in tumor suppression is extensively documented and well established, the evidence for its roles in immunity did not start to accumulate until recently. In this review, we will focus on the newly discovered functions of PTEN in the regulation of innate and adaptive immunity, including antiviral responses. Cellular & Molecular Immunology (2017) 14, 581–589; doi:10.1038/cmi.2017.30; published online 26 June 2017 Keywords: adaptive immunity; innate immunity; PTEN; tumor suppressor PTEN AND TUMOR SUPPRESSION found in tumor cells (http://cancer.sanger.ac.uk/cosmic/search? In 1997, two groups independently found that there is a tumor q = pten). Some mutations can even directly change the suppressor gene located on chromosome 10q23.1,2 Li et al.1 conformation of PTEN or can impair its phosphatase activity. named the gene PTEN, abbreviated from phosphatase and Researchers have also discovered that mutation of one of the tensin homolog (PTEN) deleted on chromosome 10, which PTEN alleles could affect the function of PTEN. -

Hypomethylation-Linked Activation of PLCE1 Impedes
Published OnlineFirst February 17, 2020; DOI: 10.1158/0008-5472.CAN-19-1912 CANCER RESEARCH | MOLECULAR CELL BIOLOGY Hypomethylation-Linked Activation of PLCE1 Impedes Autophagy and Promotes Tumorigenesis through MDM2-Mediated Ubiquitination and Destabilization of p53 Yunzhao Chen1,2, Huahua Xin1, Hao Peng1, Qi Shi1, Menglu Li1,JieYu2, Yanxia Tian1, Xueping Han1, Xi Chen1, Yi Zheng3,JunLi4, Zhihao Yang1, Lan Yang1, Jianming Hu1, Xuan Huang5, Zheng Liu5, Xiaoxi Huang5, Hong Zhou6, Xiaobin Cui1, and Feng Li1,5 ABSTRACT ◥ Esophageal squamous cell carcinoma (ESCC) is one of the dead- Significance: These findings identify hypomethylation- liest malignant diseases. Multiple studies with large clinic-based mediated activation of PLCE1 as a potential oncogene that cohorts have revealed that variations of phospholipase C epsilon 1 blocks cellular autophagy of esophageal carcinoma by facilitat- (PLCE1) correlate with esophageal cancer susceptibility. However, ing the MDM2-dependent ubiquitination of p53 and subsequent the causative role of PLCE1 in ESCC has remained elusive. Here, we degradation. observed that hypomethylation-mediated upregulation of PLCE1 Graphical Abstract: http://cancerres.aacrjournals.org/content/ expression was implicated in esophageal carcinogenesis and poor canres/80/11/2175/F1.large.jpg. prognosis in ESCC cohorts. PLCE1 inhibited cell autophagy and suppressed the protein expression of p53 and various p53-targeted genes in ESCC. Moreover, PLCE1 decreased the half-life of p53 and Normal cells Cancer cells promoted p53 ubiquitination, whereas it increased the half-life of PLCE1 Cytoplasm Cytoplasm mouse double minute 2 homolog (MDM2) and inhibited its ubiqui- wtp53 wtp53 tination, leading to MDM2 stabilization. Mechanistically, the func- MDM2 MDM2 wtp53 MDM2 MDM2 Nucleus tion of PLCE1 correlated with its direct binding to both p53 and Nucleus wtp53 Ub Ub MDM2, which promoted MDM2-dependent ubiquitination of p53 PLCE1 MDM2 wtp53 Ub wtp53 wtp53 and subsequent degradation in vitro.