Joosten, Han M.L.J.; Herst, Patricia M.; Drift, Chris Van Der
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Amino Acid Disorders 105
AMINO ACID DISORDERS 105 Massaro, A. S. (1995). Trypanosomiasis. In Guide to Clinical tions in biological fluids relatively easy. These Neurology (J. P. Mohrand and J. C. Gautier, Eds.), pp. 663– analyzers separate amino acids either by ion-ex- 667. Churchill Livingstone, New York. Nussenzweig, V., Sonntag, R., Biancalana, A., et al. (1953). Ac¸a˜o change chromatography or by high-pressure liquid de corantes tri-fenil-metaˆnicos sobre o Trypanosoma cruzi in chromatography. The results are plotted as a graph vitro: Emprego da violeta de genciana na profilaxia da (Fig. 1). The concentration of each amino acid can transmissa˜o da mole´stia de chagas por transfusa˜o de sangue. then be calculated from the size of the corresponding O Hospital (Rio de Janeiro) 44, 731–744. peak on the graph. Pagano, M. A., Segura, M. J., DiLorenzo, G. A., et al. (1999). Cerebral tumor-like American trypanosomiasis in Most amino acid disorders can be diagnosed by acquired immunodeficiency syndrome. Ann. Neurol. 45, measuring the concentrations of amino acids in 403–406. blood plasma; however, some disorders of amino Rassi, A., Trancesi, J., and Tranchesi, B. (1982). Doenc¸ade acid transport are more easily recognized through the Chagas. In Doenc¸as Infecciosas e Parasita´rias (R. Veroesi, Ed.), analysis of urine amino acids. Therefore, screening 7th ed., pp. 674–712. Guanabara Koogan, Sa˜o Paulo, Brazil. Spina-Franc¸a, A., and Mattosinho-Franc¸a, L. C. (1988). for amino acid disorders is best done using both South American trypanosomiasis (Chagas’ disease). In blood and urine specimens. Occasionally, analysis of Handbook of Clinical Neurology (P. -

Amino Acid Catabolism in Staphylococcus Aureus
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Fall 12-16-2016 Amino Acid Catabolism in Staphylococcus aureus Cortney Halsey University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Bacteriology Commons Recommended Citation Halsey, Cortney, "Amino Acid Catabolism in Staphylococcus aureus" (2016). Theses & Dissertations. 160. https://digitalcommons.unmc.edu/etd/160 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. Amino Acid Catabolism in Staphylococcus aureus By Cortney R. Halsey A DISSERTATION Presented to the Faculty of The Graduate College in the University of Nebraska In Partial Fulfillment of the Requirements For the Degree of Doctor of Philosophy Pathology and Microbiology Under the Supervision of Dr. Paul D. Fey University of Nebraska Medical Center Omaha, Nebraska October 2016 Supervisory Committee: Kenneth Bayles, Ph.D. Steven Carson, Ph.D. Paul Dunman, Ph.D. Rakesh Singh, Ph.D. ii Acknowledgements First and foremost, I would like to thank my mentor, Dr. Paul Fey, whose patience and support over the past six years has been critical to my success as a graduate student. Paul has given me opportunities to grow as a scientist and person, for which I will be forever thankful. I would also like to thank Dr. Ken Bayles, Dr. Steven Carson, Dr. Paul Dunman, and Dr. Rakesh Singh for serving on my supervisory committee. -

BIOCHEMISTRY of TRYPTOPHAN in HEALTH and DISEASE Contents
Molec. Aspects Med. Vol. 6, pp. 101-197, 1982 0098-2997/82/020101-97548.50/0 Printed in Great Britain. All rights reserved. Copyright © Pergamon Press Ltd. BIOCHEMISTRY OF TRYPTOPHAN IN HEALTH AND DISEASE David A. Bender Courtauld Institute of Biochemistry, The Middlesex Hospital Medical School, London WIP 7PN, U.K. Contents Chapter 1 THE DISCOVERY OF TRYPTOPHAN, ITS PHYSIOLOGICAL SIGNIFICANCE AND METABOLIC FATES 103 Tryptophan and glucose metabolism 105 Xanthurenic acid and insulin 105 The glucose tolerance factor 106 Inhibition of gluconeogenesis by tryptophan metabolites i07 Metabolic fates of tryptophan 108 Protein synthesis 108 Oxidative metabolism Ii0 5-Hydroxyindole synthesis 111 Intestinal bacterial metabolism iii Chapter 2 THE 5-HYDROXYINDOLE PATHWAY OF TRYPTOPHAN METABOLISM; SEROTONIN AND OTHER CENTRALLY ACTIVE TRYPTOPHAN METABOLITES 112 Tryptophan 5-hydroxylase 112 Inhibition of tryptophan hydroxylase and the carcinoid syndrome 116 Aromatic amino acid decarboxylase 118 The specificity of aromatic amino acid decarboxylase 120 Tryptophan metabolism in the pineal gland 121 Monoamine oxidase 124 The uptake of tryptophan into the brain 124 The binding of tryptophan to serum albumin 127 Competition for uptake by other neutral amino acids 129 Changes in tryptophan metabolism in response to food intake 129 Tryptophan uptake into the brain in liver failure 131 Sleep and tryptophan metabolism 134 101 102 D.A. Bender Tryptophan and serotonin in psychiatric disorders 135 Affective disorders 136 Evidence for a deficit of serotonin or tryptophan -

Histidinaemia in Mouse and Man
Arch Dis Child: first published as 10.1136/adc.49.7.545 on 1 July 1974. Downloaded from Archives of Disease in Childhood, 1974, 49, 545. Histidinaemia in mouse and man GRAHAME BULFIELD and HENRIK KACSER From the Department of Genetics, University of Edinburgh Bulfield, G., and Kacser, H. (1974). Archives of Disease in Childhood, 49, 545. Histidinaemia in mouse and man. A recently discovered mutant in the mouse was found to have very low levels of histidase. It is an autosomal recessive. In its enzymic and metabolic properties it appears to be a homologue ofhuman histidinaemia. While the homozygous mouse mutants show no overt abnormalities, offspring of histidinaemic mothers display a balance defect resulting in circling behaviour. This is associated with vestibular damage during in utero development. Mental retardation caused by human maternal phenylketonuria may have a similar aetiology. There are well recognized advantages in studying histidine and its derivatives. The enzymic and animal models in the investigation of human metabolic changes in both mouse and human diseases. The increasing number of disorders in mutants appear to be the same. In addition, man which are reported to have a genetic basis maternal histidinaemia in the mouse has a terato- (McKusick, 1971; Raine, 1972) makes it desirable to genic effect on the developing inner ear which results find and investigate animal homologues with the in abnormal behaviour of the affected offspring. same genic lesions. The genetically most in- These findings may be relevant to the aetiology of vestigated laboratory mammal is the mouse, with human histidinaemia. over 500 known mutants (Green, 1966; Searle, 1973). -

Inborn Errors of Metabolism in Adults: a Diagnostic Approach to Neurological and Psychiatric Presentations
71 2 Inborn Errors of Metabolism in Adults: A Diagnostic Approach to Neurological and Psychiatric Presentations Fanny Mochel, Frédéric Sedel 2.1 Differences Between Paediatric and Adult Phenotypes – 72 2.2 General Approach to IEM In Adulthood – 72 2.3 Specific Approaches to Neuro metabolic Presentations in Adults – 76 References – 89 J.-M. Saudubray et al. (Eds.), Inborn Metabolic Diseases, DOI 10.1007/978-3-662-49771-5_2, © Springer-Verlag Berlin Heidelberg 2016 72 Chapter 2 · Inborn Errors of Metabolism in Adults: A Diagnostic Approach to Neurological and Psychiatric Presentations Late-onset forms of IEM presenting initially in adulthood are of- In addition, some clinical signs are highly suggestive of a ten unrecognised, so that their exact prevalence is unknown [1]. particular IEM or of a particular group of IEM. Some of these Most often they have psychiatric or neurological manifestations, ‘red flags’ are listed in . Table 2.2. 2 including atypical psychosis or depression, unexplained coma, Unfortunately, in many circumstances, highly specific peripheral neuropathy, cerebellar ataxia, spastic paraparesis, de- signs or symptoms are lacking and the presentation is that of mentia, movement disorders and epilepsy [2][3][4][5]. a less specific neurological or psychiatric disorder (epilepsy, Physicians caring for adult patients with IEM are also involved in cognitive decline, psychiatric signs). In such situations, the the management of those with early onset forms who reach diagnostic approach is based on the type of clinical signs, their adulthood. The transfer of such patients from paediatric to adult clinical course (acute, acute-relapsing, with diurnal variations, care raises a number of medical, dietetic and social concerns. -

Structure and Function of Metallohydrolases in the Arginase- Deacetylase Family
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2016 Structure and Function of Metallohydrolases in the Arginase- Deacetylase Family Yang Hai University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Biochemistry Commons Recommended Citation Hai, Yang, "Structure and Function of Metallohydrolases in the Arginase-Deacetylase Family" (2016). Publicly Accessible Penn Dissertations. 1753. https://repository.upenn.edu/edissertations/1753 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/1753 For more information, please contact [email protected]. Structure and Function of Metallohydrolases in the Arginase-Deacetylase Family Abstract Arginases and deacetylases are metallohydrolases that catalyze two distinct chemical transformations. The arginases catalyze the hydrolysis of the guanidinium group of arginine by using a hydroxide ion 2+ 2+ bridging the binuclear manganese cluster (Mn A-Mn B) for nucleophilic attack. The deacetylases catalyze the hydrolysis of amide bonds by using a mononuclear Zn2+-ion activated water molecule as the nucleophile. Despite the diverse functions, metallohydrolases of the arginase-deacetylase superfamily 2+ share the same characteristic α/β hydrolase core fold and a conserved metal binding site (the Mn B site in arginase corresponds to the catalytic Zn2+ site in deacetylase) which is essential for catalysis in both enzymes. We report crystal structure of formiminoglutamase from the parasitic protozoan Trypanosoma cruzi and confirm that formiminoglutamase is a Mn2+-requiring hydrolase that belongs to the arginase- deacetylase superfamily. We also report the crystal structure of an arginase-like protein from Trypanosoma brucei (TbARG) with unknown function. Although its biological role remains enigmatic, the 2+ evolutionarily more conserved Mn B site can be readily restored in TbARG through side-directed mutagenesis. -

Endogenous Lipid Chemoattractants and Extracellular Matrix Proteins
ENDOGENOUS LIPID CHEMOATTRACTANTS AND EXTRACELLULAR MATRIX PROTEINS INVOLVED IN DEVELOPMENT OF MYXOCOCCUS XANTHUS by PATRICK DAVID CURTIS (Under the Direction of Lawrence J. Shimkets) ABSTRACT The soil bacterium Myxococcus xanthus is a model organism to study multicellular development and biofilm formation. When starved, swarms of M. xanthus cells aggregate into a multicellular architecture called a fruiting body, wherein cells differentiate into metabolically dormant myxospores. Fruiting body formation requires directed cell movement and production of an extracellular matrix (ECM) to facilitate cell-contact dependent motility (Social motility), and biofilm formation. M. xanthus displays chemotaxis towards phospholipids derived from its membrane containing the rare fatty acid 16:1ω5c. This study demonstrates that 16:1ω5c is primarily found at the sn-1 position within the major membrane phospholipid, phosphatidylethanolamine (PE), which is contrary to the established dogma of fatty acid localization in Gram-negative bacteria. Additionally, 16:1ω5c at the sn-1 position stimulates chemotaxis stronger than 16:1ω5c located at the sn-2 position. These results suggest that the endogenous lipid chemoattractants may serve as a self-recognition system. Chemotaxis towards a self-recognition marker could facilitate movement of cells into aggregation centers. Lipid chemotaxis is dependent on the ECM-associated zinc metalloprotease FibA, suggesting that the ECM may harbor protein components of extracellular signaling pathways. Protein components of prokaryotic biofilms are largely unexplored. Twenty one putative ECM-associated proteins were identified, including FibA. Many are novel proteins. A large portion of the putative ECM proteins have lipoprotein secretion signals, unusual for extracellular proteins. An MXAN4860 pilA mutant displays a 24 hour delay in fruiting body formation and sporulation compared to the pilA parent, indicating that MXAN4860 functions in the FibA-mediated developmental pathway previously described. -

Mutations in the Urocanase Gene UROC1 Are Associated with Urocanic Aciduria
1 Mutations in the urocanase gene UROC1 are associated with urocanic aciduria C Espinós, M Pineda, D Martínez-Rubio, V Lupo, A Ormazabal, M A Vilaseca, , L J M Spaapen, F Palau, R Artuch. Authors’ affiliations: Carmen Espinós, Dolores Martínez-Rubio, V Lupo, Francesc Palau, Laboratory of Genetics and Molecular Medicine, Instituto de Biomedicina, Consejo Superior de Investigaciones Científicas (CSIC), and CIBER de Enfermedades Raras (CIBERER), Valencia, Spain. Mercè Pineda, Aida Ormazabal, Maria A. Vilaseca, Rafael Artuch, Pediatric Neurology and Clinical Biochemistry Departments, Hospital Sant Joan de Déu, and CIBER de Enfermedades Raras (CIBERER), Barcelona, Spain. Leo J. M. Spaapen, Department of Biochemical Genetics, University Hospital Maastricht, The Netherlands. Corresponding author: Dr. Rafael Artuch Clinical Biochemistry Department, Hospital Sant Joan de Déu Passeig Sant Joan de Déu, 2 08950 Esplugues, Barcelona, Spain Telephone: + 34932806169 Fax: +34932803626 e-mail: [email protected] Key Words: Urocanic aciduria; urocanase gene; intermittent ataxia; mental retardation,. Wourd count: 3033 2 Key points We investigate the underlying molecular defect in a patient who suffers from urocanic aciduria presenting with mental retardation and intermittent ataxia. The biochemical findings were consistent with a defect in urocanase. We found two putative mutations in the urocanase gene UROC1 that produce two amino acid substitutions, p.L70P and p.R450C. In silico predictions, protein expression studies and enzyme activity assays suggest that none of the mutations produce a fully functional enzyme. The p.L70P substitution, which probably imply the disruption of an -helix in the N-terminus, would alter its properties and therefore, its function. The p.R450C change would render impossible any interaction between urocanase and its substrate and would loss its enzyme activity. -

Diagnose a Broad Range of Metabolic Disorders with a Single Test, Global
PEDIATRIC Assessing or diagnosing a metabolic disorder commonly requires several tests. Global Metabolomic Assisted Pathway Screen, commonly known as Global MAPS, is a unifying test GLOBAL MAPS™ for analyzing hundreds of metabolites to identify changes Global Metabolomic or irregularities in biochemical pathways. Let Global MAPS Assisted Pathway Screen guide you to an answer. Diagnose a broad range of metabolic disorders with a single test, Global MAPS Global MAPS is a large scale, semi-quantitative metabolomic profiling screen that analyzes disruptions in both individual analytes and pathways related to biochemical abnormalities. Using state-of-the-art technologies, Global Metabolomic Assisted Pathway Screen (Global MAPS) provides small molecule metabolic profiling to identify >700 metabolites in human plasma, urine, or cerebrospinal fluid. Global MAPS identifies inborn errors of metabolism (IEMs) that would ordinarily require many different tests. This test defines biochemical pathway errors not currently detected by routine clinical or genetic testing. IEMs are inherited metabolic disorders that prevent the body from converting one chemical compound to another or from transporting a compound in or out of a cell. NORMAL PROCESS METABOLIC ERROR These processes are necessary for essentially all bodily functions. Most IEMs are caused by defects in the enzymes that help process nutrients, which result in an accumulation of toxic substances or a deficiency of substances needed for normal body function. Making a swift, accurate diagnosis -
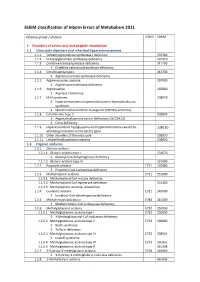
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

MARTI-ARBONA-DISSERTATION.Pdf (5.556Mb)
MECHANISTIC CHARACTERIZATION OF MEMBERS OF THE AMIDOHYDROLASE SUPERFAMILY A Dissertation by RICARDO MARTÍ ARBONA Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY December 2006 Major Subject: Chemistry MECHANISTIC CHARACTERIZATION OF MEMBERS OF THE AMIDOHYDROLASE SUPERFAMILY A Dissertation by RICARDO MARTÍ ARBONA Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Approved by: Chair of Committee, Frank M. Raushel Committee Members, Paul A. Lindahl Eric E. Simanek Jerry Tsai Head of Department, Emile A. Schweikert December 2006 Major Subject: Chemistry iii ABSTRACT Mechanistic Characterization of Members of the Amidohydrolase Superfamily. (December 2006) Ricardo Martí Arbona, B.S., Universidad de Puerto Rico, at Río Piedras Chair of Advisory Committee: Dr. Frank M. Raushel The amidohydrolase superfamily is a functionally diverse group of enzymes found in every organism sequenced to date. The landmark for this superfamily is the conservation of a (/)8-barrel structural fold. Isoaspartyl dipeptidase (IAD) from Escherichia coli catalyzes the hydrolytic cleavage of -aspartyl dipeptides. Structural studies of the wild-type enzyme demonstrate that the active site consists of a binuclear metal center. Bell-shaped pH-rate profiles are observed for all four metal-substituted forms of the wild-type enzyme and the site-directed mutants, E77Q and Y137F. Structural analysis of IAD with the bound substrate and site-directed mutagenesis shows the importance of the side chains of residues Glu-77, Tyr-137, Arg-169, Arg-233, Asp- 285, and Ser-289 in the substrate binding and hydrolysis. -

Supplemental Table S1: Comparison of the Deleted Genes in the Genome-Reduced Strains
Supplemental Table S1: Comparison of the deleted genes in the genome-reduced strains Legend 1 Locus tag according to the reference genome sequence of B. subtilis 168 (NC_000964) Genes highlighted in blue have been deleted from the respective strains Genes highlighted in green have been inserted into the indicated strain, they are present in all following strains Regions highlighted in red could not be deleted as a unit Regions highlighted in orange were not deleted in the genome-reduced strains since their deletion resulted in severe growth defects Gene BSU_number 1 Function ∆6 IIG-Bs27-47-24 PG10 PS38 dnaA BSU00010 replication initiation protein dnaN BSU00020 DNA polymerase III (beta subunit), beta clamp yaaA BSU00030 unknown recF BSU00040 repair, recombination remB BSU00050 involved in the activation of biofilm matrix biosynthetic operons gyrB BSU00060 DNA-Gyrase (subunit B) gyrA BSU00070 DNA-Gyrase (subunit A) rrnO-16S- trnO-Ala- trnO-Ile- rrnO-23S- rrnO-5S yaaC BSU00080 unknown guaB BSU00090 IMP dehydrogenase dacA BSU00100 penicillin-binding protein 5*, D-alanyl-D-alanine carboxypeptidase pdxS BSU00110 pyridoxal-5'-phosphate synthase (synthase domain) pdxT BSU00120 pyridoxal-5'-phosphate synthase (glutaminase domain) serS BSU00130 seryl-tRNA-synthetase trnSL-Ser1 dck BSU00140 deoxyadenosin/deoxycytidine kinase dgk BSU00150 deoxyguanosine kinase yaaH BSU00160 general stress protein, survival of ethanol stress, SafA-dependent spore coat yaaI BSU00170 general stress protein, similar to isochorismatase yaaJ BSU00180 tRNA specific adenosine