Pedigree and Molecular Analyses in the Assessment of Genetic Variability of the Polish Greyhound
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Systematyka Ras Na Krajową Wystawę Psów Ras Myśliwskich
SYSTEMATYKA RAS NA KRAJOWĄ WYSTAWĘ PSÓW RAS MYŚLIWSKICH GROUP III - TERRIERS / GRUPA III - TERIERY Section 1 - Large and medium sized Terriers / Sekcja 1 - Teriery duże i średnie -Terrier Brasileiro (341) Terier brazylijski - Deutscher Jagdterrier (103) Niemiecki terier myśliwski -Airedale Terrier (7) -Border Terrier (10) -Fox Terrier (Smooth) (12) Foksterier krótkowłosy -Fox Terrier (Wire) (169) Foksterier szorstkowłosy -Lakeland Terrier (70) -Manchester Terrier (71) -Parson Russell Terrier (339) -Terier walijski -Irish Glen of Imaal Terrier (302) -Irish Soft Coated Wheaten Terrier (40) -Irish Terrier (139) -Terier irlandzki Kerry Blue Terrier (3) Section 2 - Small sized Terriers / Sekcja 2 - Teriery małe -Australian Terrier (8) Terier australijski -Ceský Teriér (246) (Cesky Terrier) Terier czeski -Cairn Terrier (4) -Dandie Dinmont Terrier (168) -Jack Russell Terrier (345) -Norfolk Terrier (272) -Norwich Terrier (72) -Scottish Terrier (73) Terier szkocki -Sealyham Terrier (74) -Skye Terrier (75) -Nihon Teria (259) (Japanese Terrier) GROUP IV - DACHSHUND / GRUPA IV – JAMNIKI Dachshund (Teckel) (148) a) Dachshund (Standard) - Kurzhaar (Smooth-haired) / Jamnik krótkowłosy standardowy - Langhaar (Long-haired) / Jamnik długowłosy standardowy - Rauhhaar (Wire-haired) / Jamnik szorstkowłosy standardowy b) Zwerg-Dachshund (Miniature) - Kurzhaar (Smooth-haired) /Jamnik krótkowłosy miniaturowy - Langhaar (Long-haired) / Jamnik długowłosy miniaturowy - Rauhhaar (Wire-haired) / Jamnik szorstkowłosy miniaturowy c) Kaninchen-Dachshund (Rabbit Dachshund) -

Dog Breeds Pack 1 Professional Vector Graphics Page 1
DOG BREEDS PACK 1 PROFESSIONAL VECTOR GRAPHICS PAGE 1 Affenpinscher Afghan Hound Aidi Airedale Terrier Akbash Akita Inu Alano Español Alaskan Klee Kai Alaskan Malamute Alpine Dachsbracke American American American American Akita American Bulldog Cocker Spaniel Eskimo Dog Foxhound American American Mastiff American Pit American American Hairless Terrier Bull Terrier Staffordshire Terrier Water Spaniel Anatolian Anglo-Français Appenzeller Shepherd Dog de Petite Vénerie Sennenhund Ariege Pointer Ariegeois COPYRIGHT (c) 2013 FOLIEN.DS. ALL RIGHTS RESERVED. WWW.VECTORART.AT DOG BREEDS PACK 1 PROFESSIONAL VECTOR GRAPHICS PAGE 2 Armant Armenian Artois Hound Australian Australian Kelpie Gampr dog Cattle Dog Australian Australian Australian Stumpy Australian Terrier Austrian Black Shepherd Silky Terrier Tail Cattle Dog and Tan Hound Austrian Pinscher Azawakh Bakharwal Dog Barbet Basenji Basque Basset Artésien Basset Bleu Basset Fauve Basset Griffon Shepherd Dog Normand de Gascogne de Bretagne Vendeen, Petit Basset Griffon Bavarian Mountain Vendéen, Grand Basset Hound Hound Beagle Beagle-Harrier COPYRIGHT (c) 2013 FOLIEN.DS. ALL RIGHTS RESERVED. WWW.VECTORART.AT DOG BREEDS PACK 2 PROFESSIONAL VECTOR GRAPHICS PAGE 3 Belgian Shepherd Belgian Shepherd Bearded Collie Beauceron Bedlington Terrier (Tervuren) Dog (Groenendael) Belgian Shepherd Belgian Shepherd Bergamasco Dog (Laekenois) Dog (Malinois) Shepherd Berger Blanc Suisse Berger Picard Bernese Mountain Black and Berner Laufhund Dog Bichon Frisé Billy Tan Coonhound Black and Tan Black Norwegian -

Polish Greyhound
FEDERATION OF CYNOLOGY FOR EUROPE ФЕДЕРАЦИЯ ПО КИНОЛОГИЯ ЗА ЕВРОПА 01.10.1991/EN FCE-Standard № 10-333 POLISH GREYHOUND (Chart Polski) 2 TRANSLATION: Mrs.Peggy Davis. ORIGIN: Poland. DATE OF PUBLICATION OF THE OFFICIAL VALID STANDARD: 01.10.1999. UTILIZATION: Hunting dog not only for hare, fox, roe-deer and bustard, but also for the wolf. FCЕ-CLASSIFICATION : Group 10 Sighthounds. Section 3 Short-haired Sighthounds. Without working trial. BRIEF HISTORICAL SUMMARY: The presence of the Chart Polski in Poland is attested since the 13th century; this breed goes probably back to Asiatic sighthounds of Saluki type. The Borzoi being unknown before the reign of Iwan the Terrible during the XVIth century, it is impossible, as claimed by the Russian author Sabaniejew, that the Chart Polski would be the result of interbreeding between the Greyhound and the Borzoi. The mention of the Chart Polski in the literature, especially the hunt-literature, is frequent and the iconographic representations are noticeably unvarying. This uniform general appearance in drawings and paintings proves, that, in spite of different interbreeding, the original aspect of the breed has remained unchanged up to the end of the XIXth century. GENERAL APPEARANCE: The Polish Greyhound is a dog of great size, powerful, muscular, definitely stronger and less fine in shape than the other short-haired sighthounds (he must not, however, be heavy nor lethargic). In his appearance, he is similar to the Asiatic greyhound who is his ancestor. FCE-St. №10-333/01.01.1999 3 The strong frame, the short coupled body, the distinctly visible musculature and the powerful jaws show that this dog has been used for hunting in the difficult conditions of the Polish climate. -

Qualified for Crufts 2019
Qualified for Crufts 2019 at the Benelux Winner Show in Amsterdam 9th August 2018 Breed DogName ChipTattoo Gender Owner Australian Kelpie Yaparoos Arramagong Taz 528140000622424 NHSB 3013958 M Marielle Van Der Heeden Australian Kelpie Didaktic's Omg 978101081715689 FI 39682/17 F Marika Lehtonen Belgian Shepherd Dog, Groenendael Revloch Figo 956000008566119 IKC Z48074 M Jean Lawless Belgian Shepherd Dog, Groenendael Revloch Karma Of Woofhouse 941000017117451 IKC Z84192 F Pamela Mckenzie-Hewitt Belgian Shepherd Dog, Laekenois Ruby Rivers Tristan 968000005743426 VDH SE53174/2011 M Karl Strugalla Belgian Shepherd Dog, Malinois Alcosetha'S Jalco 528140000572448 SE 46836/2014 M Ylva Persson Belgian Shepherd Dog, Malinois Boholdt Gloria Gaynor 208250000102703 DKK 18423/2017 F Annelise Sindalsen Belgian Shepherd Dog, Tervueren Glimbi S El Ninõ V. Ginger 208210000413061 DKK DK07707/2013 M Erik Breukelman Belgian Shepherd Dog, Tervueren Ketunpolun Kiss Me Kate 978101081395539 LSVK BST0051/15 F Marciukaitiene Asta Schipperke Slavjan Al Capone 643094100436648 RKF 3977884 M Maria Spiridonova Schipperke BEACHVIEW'S TIL DEATH DO US PART 1653392 AKC NP33073101 F Lindsey @ Joanne Mclachlan Czechoslovakian Wolfdog My Kayden De La Legende Du Loup Noir 250268500976212 LOF 4004/0 M Regis Haydo Czechoslovakian Wolfdog Arimminum Yasmeen 380260010336510 ROI 15/155486 F Scaglia Laura German Shepherd Dog, double coat Omero Haus Dando 804098100057016 UKU 0182383 M Kholodna Ievgeniia German Shepherd Dog, double coat Adele Da Soc Port 620098100741122 LOP 507637 F -

Canis-Adn-Mix Razas Caninas
CANIS-ADN-MIX RAZAS CANINAS Affenpinscher Bohemian Shepherd Afghan Hound Bolognese Airedale Terrier Border Collie Akita Border Terrier Alaskan Klee Kai Borzoi Alaskan Malamute Boston Terrier Alpine Dachsbracke Bouvier Des Flanders American English Coonhound Boxer American Eskimo Dog Boykin Spaniel American Foxhound Bracco Italiano American Hairless Rat Terrier Braque du Bourbonnais American Indian Dog Brazilian Terrier American Staffordshire Terrier Briard American Water Spaniel Brussels Griffon Anatolian Shepherd Dog Bulgarian Shepherd Appenzell Cattle Dog Bull Arab Argentine Dogo Bull Terrier (Miniature) Australian Cattle Dog Bull Terrier (Standard) Australian Kelpie Bulldog (American) Australian Labradoodle Bulldog (Standard) Australian Shepherd Bullmastiff Australian Stumpy Tail Cattle Dog Cairn Terrier Australian Terrier Canaan Dog Azawakh Canadian Eskimo Dog Barbet Cane Corso Basenji Cardigan Welsh Corgi Basset Bleu de Gascogne Catahoula Leopard Dog Basset Fauve de Bretagne Catalan Sheepdog Basset Griffon Vendeen (Grand) Caucasian Shepherd Dog Basset Griffon Vendeen (Petit) Cavalier King Charles Spaniel Basset Hound Central Asian Ovcharka Bavarian Mountain Hound Cesky Terrier Beagle Chesapeake Bay Retriever Bearded Collie Chihuahua Beauceron Chinese Crested Bedlington Terrier Chinese Shar-Pei Belgian Malinois Chinook Belgian Mastiff Chow Chow Belgian Sheepdog Cirneco del Etna Belgian Tervuren Clumber Spaniel Bergamasco Cocker Spaniel Berger Picard Collie Bernese Mountain Dog Continental Toy Spaniel (phalene and papillon) Bichon -
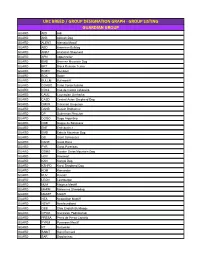
Ukc Breed / Group Designation Graph
UKC BREED / GROUP DESIGNATION GRAPH - GROUP LISTING GUARDIAN GROUP GUARD AIDI Aidi GUARD AKB Akbash Dog GUARD ALENT Alentejo Mastiff GUARD ABD American Bulldog GUARD ANAT Anatolian Shepherd GUARD APN Appenzeller GUARD BMD Bernese Mountain Dog GUARD BRT Black Russian Terrier GUARD BOER Boerboel GUARD BOX Boxer GUARD BULLM Bullmastiff GUARD CORSO Cane Corso Italiano GUARD CDCL Cao de Castro Laboreiro GUARD CAUC Caucasian Ovcharka GUARD CASD Central Asian Shepherd Dog GUARD CMUR Cimarron Uruguayo GUARD DANB Danish Broholmer GUARD DP Doberman Pinscher GUARD DOGO Dogo Argentino GUARD DDB Dogue de Bordeaux GUARD ENT Entlebucher GUARD EMD Estrela Mountain Dog GUARD GS Giant Schnauzer GUARD DANE Great Dane GUARD PYR Great Pyrenees GUARD GSMD Greater Swiss Mountain Dog GUARD HOV Hovawart GUARD KAN Kangal Dog GUARD KSHPD Karst Shepherd Dog GUARD KOM Komondor GUARD KUV Kuvasz GUARD LEON Leonberger GUARD MJM Majorca Mastiff GUARD MARM Maremma Sheepdog GUARD MASTF Mastiff GUARD NEA Neapolitan Mastiff GUARD NEWF Newfoundland GUARD OEB Olde English Bulldogge GUARD OPOD Owczarek Podhalanski GUARD PRESA Perro de Presa Canario GUARD PYRM Pyrenean Mastiff GUARD RT Rottweiler GUARD SAINT Saint Bernard GUARD SAR Sarplaninac GUARD SC Slovak Cuvac GUARD SMAST Spanish Mastiff GUARD SSCH Standard Schnauzer GUARD TM Tibetan Mastiff GUARD TJAK Tornjak GUARD TOSA Tosa Ken SCENTHOUND GROUP SCENT AD Alpine Dachsbracke SCENT B&T American Black & Tan Coonhound SCENT AF American Foxhound SCENT ALH American Leopard Hound SCENT AFVP Anglo-Francais de Petite Venerie SCENT -

Chart Polski (Polish Greyhound) Official UKC Breed Standard Sighthound and Pariah Dog Group ©Copyright 1996, United Kennel Club
Chart Polski (Polish Greyhound) Official UKC Breed Standard Sighthound and Pariah Dog Group ©Copyright 1996, United Kennel Club large and strong, he must never be overdone to the point of heaviness or lethargy. CHARACTERISTICS The Chart Polski is self assured, confident, reserved and brave. He is a fast, skillful and untiring hunter and has a lively and penetrating gaze. HEAD The head is long, lean and strong, with the muzzle as long as, or slightly longer than, the skull, and their top lines slightly divergent in profile. The skull and muzzle blend cleanly together at the sides of the head and there is a slight stop. SKULL - The upper part of the skull is flat. There is a The goals and purposes of this breed standard include: slight frontal furrow, and the frontal bones and to furnish guidelines for breeders who wish to maintain eyebrows are lightly marked. the quality of their breed and to improve it; to advance MUZZLE - The muzzle is very strong in front of the eyes. this breed to a state of similarity throughout the world; It tapers slightly towards the tip, but in no way appears and to act as a guide for judges. to be pointed; instead it is rather blunt for a Breeders and judges have the responsibility to avoid sighthound. The position of the nose is somewhat any conditions or exaggerations that are detrimental to below the upper line of the muzzle. The lips are well the health, welfare, essence and soundness of this defined, but clean and never pendulous. They do not breed, and must take the responsibility to see that cover the lower jaw. -

Kurzform Rasse / Abbreviation Breed Stand/As Of: Sept
Kurzform Rasse / Abbreviation Breed Stand/as of: Sept. 2016 A AP Affenpinscher / Monkey Terrier AH Afghanischer Windhund / Afgan Hound AID Atlas Berghund / Atlas Mountain Dog (Aidi) AT Airedale Terrier AK Akita AM Alaskan Malamute DBR Alpenländische Dachsbracke / Alpine Basset Hound AA American Akita ACS American Cocker Spaniel / American Cocker Spaniel AFH Amerikanischer Fuchshund / American Foxhound AST American Staffordshire Terrier AWS Amerikanischer Wasserspaniel / American Water Spaniel AFPV Small French English Hound (Anglo-francais de petite venerie) APPS Appenzeller Sennenhund / Appenzell Mountain Dog ARIE Ariegeois / Arigie Hound ACD Australischer Treibhund / Australian Cattledog KELP Australian Kelpie ASH Australian Shepherd SILT Australian Silky Terrier STCD Australian Stumpy Tail Cattle Dog AUST Australischer Terrier / Australian Terrier AZ Azawakh B BARB Franzosicher Wasserhund / French Water Dog (Barbet) BAR Barsoi / Russian Wolfhound (Borzoi) BAJI Basenji BAN Basst Artesien Normad / Norman Artesien Basset (Basset artesien normand) BBG Blauer Basset der Gascogne / Bue Cascony Basset (Basset bleu de Gascogne) BFB Tawny Brittany Basset (Basset fauve de Bretagne) BASH Basset Hound BGS Bayrischer Gebirgsschweisshund / Bavarian Mountain Hound BG Beagle BH Beagle Harrier BC Bearded Collie BET Bedlington Terrier BBS Weisser Schweizer Schäferhund / White Swiss Shepherd Dog (Berger Blanc Suisse) BBC Beauceron (Berger de Beauce) BBR Briard (Berger de Brie) BPIC Picardieschäferhund / Picardy Shepdog (Berger de Picardie (Berger Picard)) -

Breed Specific Instructions (BSI) Regarding Exaggerations in Pedigree Dogs
By the Nordic Kennel Clubs 2014 Applicable from 2014 Breed Specific Instructions (BSI) regarding exaggerations in pedigree dogs DANSK KENNEL KLUB HUNDARÆKTARFÉLAG ÍSLANDS NORSK KENNEL KLUB SUOMEN KENNELLIITTO SVENSKA KENNELKLUBBEN Index Introduction .............................................................................................................................4 Application ..............................................................................................................................6 Basics for all dogs ...................................................................................................................8 Breed types ...........................................................................................................................10 FCI GROUP 1 Sheepdogs and Cattle Dogs ..............................................................................12 FCI GROUP 2 Pinscher and Schnauzer - Molossoïd Breeds - Swiss Mountain and Cattle Dogs ...14 FCI GROUP 3 Terriers .............................................................................................................19 FCI GROUP 4 Dachshunds ......................................................................................................21 FCI GROUP 5 Spitz and Primitive types ...................................................................................22 FCI GROUP 6 Scenthounds and Related Breeds ......................................................................24 FCI GROUP 7 Pointing Dogs ....................................................................................................26 -

Benelux Winner - Amsterdam 9Th August 2018 FCI Gr
Benelux Winner - Amsterdam 9th August 2018 FCI Gr. Breed Name of the Dog Pedigree Gender Date of Birth Owner 1 Australian Kelpie Yaparoos Arramagong Taz NHSB 3013958 M 29-07-2015 Marielle Van Der Heeden 1 Australian Kelpie Fly Samba Fly Di Casa Hesselink ROI 15/3530 F 23-10-2014 Eleonora Marinelli 1 Belgian Shepherd Dog, Groenendael Revloch Figo IKC Z48074 M 19-11-2012 Jean Lawless 1 Belgian Shepherd Dog, Groenendael Revloch Karma Of Woofhouse IKC Z84192 F 25-09-2014 Pamela Mckenzie-Hewitt 1 Belgian Shepherd Dog, Laekenois Ruby Rivers Tristan VDH SE53174/2011 M 26-08-2011 Karl Strugalla 1 Belgian Shepherd Dog, Malinois Alcosetha'S Jalco SE 46836/2014 M 17-04-2014 Ylva Persson 1 Belgian Shepherd Dog, Tervueren Glimbi S El Ninõ V. Ginger DKK DK07707/2013 M 9-04-2013 Erik Breukelman 1 Belgian Shepherd Dog, Tervueren Ketunpolun Kiss Me Kate LSVK BST0051/15 F 28-05-2015 Marciukaitiene Asta 1 Schipperke Slavjan Al Capone RKF 3977884 M 29-06-2014 Maria Spiridonova 1 Schipperke BEACHVIEW'S TIL DEATH DO US PART AKC NP33073101 F 22-03-2012 Lindsey @ Joanne Mclachlan 1 Czechoslovakian Wolfdog My Kayden De La Legende Du Loup Noir LOF 4004/0 M 7-01-2016 Regis Haydo 1 Czechoslovakian Wolfdog Arimminum Yasmeen ROI 15/155486 F 6-08-2015 Scaglia Laura 1 German Shepherd Dog, double coat Omero Haus Dando UKU 0182383 M 28-08-2013 Kholodna Ievgeniia 1 German Shepherd Dog, double coat Adele Da Soc Port LOP 507637 F 2-08-2014 Alexandre Chaves 1 German Shepherd Dog, long and harsh outer coat Aurora Haus Dando UKU 0237200 F 20-02-2015 Maryna Antonova 1 Berger -

HSVMA Guide to Congenital and Heritable Disorders in Dogs
GUIDE TO CONGENITAL AND HERITABLE DISORDERS IN DOGS Includes Genetic Predisposition to Diseases Special thanks to W. Jean Dodds, D.V.M. for researching and compiling the information contained in this guide. Dr. Dodds is a world-renowned vaccine research scientist with expertise in hematology, immunology, endocrinology and nutrition. Published by The Humane Society Veterinary Medical Association P.O. Box 208, Davis, CA 95617, Phone: 530-759-8106; Fax: 530-759-8116 First printing: August 1994, revised August 1997, November 2000, January 2004, March 2006, and May 2011. Introduction: Purebred dogs of many breeds and even mixed breed dogs are prone to specific abnormalities which may be familial or genetic in nature. Often, these health problems are unapparent to the average person and can only be detected with veterinary medical screening. This booklet is intended to provide information about the potential health problems associated with various purebred dogs. Directory Section I A list of 182 more commonly known purebred dog breeds, each of which is accompanied by a number or series of numbers that correspond to the congenital and heritable diseases identified and described in Section II. Section II An alphabetical listing of congenital and genetically transmitted diseases that occur in purebred dogs. Each disease is assigned an identification number, and some diseases are followed by the names of the breeds known to be subject to those diseases. How to use this book: Refer to Section I to find the congenital and genetically transmitted diseases associated with a breed or breeds in which you are interested. Refer to Section II to find the names and definitions of those diseases. -
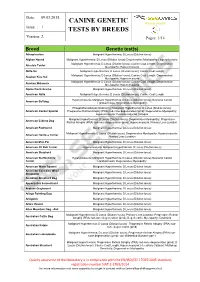
Canine Genetic Tests by Breeds
Date: 09.03.2015. CANINE GENETIC Animal Health Issue : 1. TESTS BY BREEDS Version: 2. Pages: 1/14 Breed Genetic test(s) Affenpinscher Malignant Hyperthermia; D Locus (Dilution locus) Afghan Hound Malignant Hyperthermia; D Locus (Dilution locus); Degenerative Myelopathy; Hyperuricosuria Malignant Hyperthermia; D Locus (Dilution locus); Canine Coat Length; Degenerative Airedale Terrier Myelopathy; Hyperuricosuria Akita-Inu Malignant Hyperthermia; D Locus (Dilution locus); Canine Coat Length Malignant Hyperthermia; D Locus (Dilution locus); Canine Coat Length; Degenerative Alaskan Klee Kai Myelopathy; Hyperuricosuria Malignant Hyperthermia; D Locus (Dilution locus); Canine Coat Length; Degenerative Alaskan Malamute Myelopathy; Hyperuricosuria Alpine Dachsbracke Malignant Hyperthermia; D Locus (Dilution locus) American Akita Malignant Hyperthermia; D Locus (Dilution locus); Canine Coat Length Hyperuricosuria; Malignant Hyperthermia; D Locus (Dilution locus); Neuronal Ceroid American Bulldog Lipofuscinosis; Degenerative Myelopathy Phosphofructokinase Deficiency; Malignant Hyperthermia; D Locus (Dilution locus); American Cocker Spaniel Progressive Retinal Atrophy (PRA) rod-cone degeneration (prcd); Degenerative Myelopathy; Hyperuricosuria; Exercise-induced Collapse Malignant Hyperthermia; D Locus (Dilution locus); Degenerative Myelopathy; Progressive American Eskimo Dog Retinal Atrophy (PRA) rod-cone degeneration (prcd); Hyperuricosuria; Primary Lens Luxation American Foxhound Malignant Hyperthermia; D Locus (Dilution locus) Malignant Hyperthermia;