Contribution of the Microbial Communities Detected on an Oil Painting on Canvas to Its Biodeterioration
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Desulfuribacillus Alkaliarsenatis Gen. Nov. Sp. Nov., a Deep-Lineage
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central Extremophiles (2012) 16:597–605 DOI 10.1007/s00792-012-0459-7 ORIGINAL PAPER Desulfuribacillus alkaliarsenatis gen. nov. sp. nov., a deep-lineage, obligately anaerobic, dissimilatory sulfur and arsenate-reducing, haloalkaliphilic representative of the order Bacillales from soda lakes D. Y. Sorokin • T. P. Tourova • M. V. Sukhacheva • G. Muyzer Received: 10 February 2012 / Accepted: 3 May 2012 / Published online: 24 May 2012 Ó The Author(s) 2012. This article is published with open access at Springerlink.com Abstract An anaerobic enrichment culture inoculated possible within a pH range from 9 to 10.5 (optimum at pH with a sample of sediments from soda lakes of the Kulunda 10) and a salt concentration at pH 10 from 0.2 to 2 M total Steppe with elemental sulfur as electron acceptor and for- Na? (optimum at 0.6 M). According to the phylogenetic mate as electron donor at pH 10 and moderate salinity analysis, strain AHT28 represents a deep independent inoculated with sediments from soda lakes in Kulunda lineage within the order Bacillales with a maximum of Steppe (Altai, Russia) resulted in the domination of a 90 % 16S rRNA gene similarity to its closest cultured Gram-positive, spore-forming bacterium strain AHT28. representatives. On the basis of its distinct phenotype and The isolate is an obligate anaerobe capable of respiratory phylogeny, the novel haloalkaliphilic anaerobe is suggested growth using elemental sulfur, thiosulfate (incomplete as a new genus and species, Desulfuribacillus alkaliar- T T reduction) and arsenate as electron acceptor with H2, for- senatis (type strain AHT28 = DSM24608 = UNIQEM mate, pyruvate and lactate as electron donor. -

Numidum Massiliense Gen. Nov., Sp. Nov., a New Member of the Bacillaceae Family Isolated from the Human Gut
Accepted Manuscript Numidum massiliense gen. nov., sp. nov., a new member of the Bacillaceae family isolated from the human gut Maryam Tidjani Alou, Thi-Tien Nguyen, Nicholas Armstrong, Jaishriram Rathored, Saber Khelaifia, Didier Raoult, Pierre-Edouard Fournier, Jean-Christophe Lagier PII: S2052-2975(16)30042-7 DOI: 10.1016/j.nmni.2016.05.009 Reference: NMNI 175 To appear in: New Microbes and New Infections Received Date: 15 April 2016 Revised Date: 10 May 2016 Accepted Date: 12 May 2016 Please cite this article as: Alou MT, Nguyen T-T, Armstrong N, Rathored J, Khelaifia S, Raoult D, Fournier P-E, Lagier J-C, Numidum massiliense gen. nov., sp. nov., a new member of the Bacillaceae family isolated from the human gut, New Microbes and New Infections (2016), doi: 10.1016/ j.nmni.2016.05.009. This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. ACCEPTED MANUSCRIPT Numidum massiliense gen. nov., sp. nov., a new member of the Bacillaceae family isolated from the human gut Maryam Tidjani Alou 1, Thi-Tien Nguyen 1, Nicholas Armstrong 1, Jaishriram Rathored 1, Saber Khelaifia 1, Didier Raoult 1,2 , Pierre-Edouard Fournier 1, and Jean-Christophe Lagier 1.* 1Aix-Marseille Université, URMITE, UM63, CNRS7278, IRD198, Inserm 1095, Faculté de médecine, 27 Boulevard jean Moulin, 13385 Marseille cedex 05, France. -

Bacillus Coagulans S-Lac and Bacillus Subtilis TO-A JPC, Two Phylogenetically Distinct Probiotics
RESEARCH ARTICLE Complete Genomes of Bacillus coagulans S-lac and Bacillus subtilis TO-A JPC, Two Phylogenetically Distinct Probiotics Indu Khatri☯, Shailza Sharma☯, T. N. C. Ramya*, Srikrishna Subramanian* CSIR-Institute of Microbial Technology, Sector 39A, Chandigarh, India ☯ These authors contributed equally to this work. * [email protected] (TNCR); [email protected] (SS) a11111 Abstract Several spore-forming strains of Bacillus are marketed as probiotics due to their ability to survive harsh gastrointestinal conditions and confer health benefits to the host. We report OPEN ACCESS the complete genomes of two commercially available probiotics, Bacillus coagulans S-lac Citation: Khatri I, Sharma S, Ramya TNC, and Bacillus subtilis TO-A JPC, and compare them with the genomes of other Bacillus and Subramanian S (2016) Complete Genomes of Lactobacillus. The taxonomic position of both organisms was established with a maximum- Bacillus coagulans S-lac and Bacillus subtilis TO-A likelihood tree based on twenty six housekeeping proteins. Analysis of all probiotic strains JPC, Two Phylogenetically Distinct Probiotics. PLoS of Bacillus and Lactobacillus reveal that the essential sporulation proteins are conserved in ONE 11(6): e0156745. doi:10.1371/journal. pone.0156745 all Bacillus probiotic strains while they are absent in Lactobacillus spp. We identified various antibiotic resistance, stress-related, and adhesion-related domains in these organisms, Editor: Niyaz Ahmed, University of Hyderabad, INDIA which likely provide support in exerting probiotic action by enabling adhesion to host epithe- lial cells and survival during antibiotic treatment and harsh conditions. Received: March 15, 2016 Accepted: May 18, 2016 Published: June 3, 2016 Copyright: © 2016 Khatri et al. -

Thèses Traditionnelles
UNIVERSITÉ D’AIX-MARSEILLE FACULTÉ DE MÉDECINE DE MARSEILLE ECOLE DOCTORALE DES SCIENCES DE LA VIE ET DE LA SANTÉ THÈSE Présentée et publiquement soutenue devant LA FACULTÉ DE MÉDECINE DE MARSEILLE Le 23 Novembre 2017 Par El Hadji SECK Étude de la diversité des procaryotes halophiles du tube digestif par approche de culture Pour obtenir le grade de DOCTORAT d’AIX-MARSEILLE UNIVERSITÉ Spécialité : Pathologie Humaine Membres du Jury de la Thèse : Mr le Professeur Jean-Christophe Lagier Président du jury Mr le Professeur Antoine Andremont Rapporteur Mr le Professeur Raymond Ruimy Rapporteur Mr le Professeur Didier Raoult Directeur de thèse Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes, UMR 7278 Directeur : Pr. Didier Raoult 1 Avant-propos : Le format de présentation de cette thèse correspond à une recommandation de la spécialité Maladies Infectieuses et Microbiologie, à l’intérieur du Master des Sciences de la Vie et de la Santé qui dépend de l’Ecole Doctorale des Sciences de la Vie de Marseille. Le candidat est amené à respecter des règles qui lui sont imposées et qui comportent un format de thèse utilisé dans le Nord de l’Europe et qui permet un meilleur rangement que les thèses traditionnelles. Par ailleurs, la partie introduction et bibliographie est remplacée par une revue envoyée dans un journal afin de permettre une évaluation extérieure de la qualité de la revue et de permettre à l’étudiant de commencer le plus tôt possible une bibliographie exhaustive sur le domaine de cette thèse. Par ailleurs, la thèse est présentée sur article publié, accepté ou soumis associé d’un bref commentaire donnant le sens général du travail. -

Thermolongibacillus Cihan Et Al
Genus Firmicutes/Bacilli/Bacillales/Bacillaceae/ Thermolongibacillus Cihan et al. (2014)VP .......................................................................................................................................................................................... Arzu Coleri Cihan, Department of Biology, Faculty of Science, Ankara University, Ankara, Turkey Kivanc Bilecen and Cumhur Cokmus, Department of Molecular Biology & Genetics, Faculty of Agriculture & Natural Sciences, Konya Food & Agriculture University, Konya, Turkey Ther.mo.lon.gi.ba.cil’lus. Gr. adj. thermos hot; L. adj. Type species: Thermolongibacillus altinsuensis E265T, longus long; L. dim. n. bacillus small rod; N.L. masc. n. DSM 24979T, NCIMB 14850T Cihan et al. (2014)VP. .................................................................................. Thermolongibacillus long thermophilic rod. Thermolongibacillus is a genus in the phylum Fir- Gram-positive, motile rods, occurring singly, in pairs, or micutes,classBacilli, order Bacillales, and the family in long straight or slightly curved chains. Moderate alka- Bacillaceae. There are two species in the genus Thermo- lophile, growing in a pH range of 5.0–11.0; thermophile, longibacillus, T. altinsuensis and T. kozakliensis, isolated growing in a temperature range of 40–70∘C; halophile, from sediment and soil samples in different ther- tolerating up to 5.0% (w/v) NaCl. Catalase-weakly positive, mal hot springs, respectively. Members of this genus chemoorganotroph, grow aerobically, but not under anaer- are thermophilic (40–70∘C), halophilic (0–5.0% obic conditions. Young cells are 0.6–1.1 μm in width and NaCl), alkalophilic (pH 5.0–11.0), endospore form- 3.0–8.0 μm in length; cells in stationary and death phases ing, Gram-positive, aerobic, motile, straight rods. are 0.6–1.2 μm in width and 9.0–35.0 μm in length. -

Appendix 1. Validly Published Names, Conserved and Rejected Names, And
Appendix 1. Validly published names, conserved and rejected names, and taxonomic opinions cited in the International Journal of Systematic and Evolutionary Microbiology since publication of Volume 2 of the Second Edition of the Systematics* JEAN P. EUZÉBY New phyla Alteromonadales Bowman and McMeekin 2005, 2235VP – Valid publication: Validation List no. 106 – Effective publication: Names above the rank of class are not covered by the Rules of Bowman and McMeekin (2005) the Bacteriological Code (1990 Revision), and the names of phyla are not to be regarded as having been validly published. These Anaerolineales Yamada et al. 2006, 1338VP names are listed for completeness. Bdellovibrionales Garrity et al. 2006, 1VP – Valid publication: Lentisphaerae Cho et al. 2004 – Valid publication: Validation List Validation List no. 107 – Effective publication: Garrity et al. no. 98 – Effective publication: J.C. Cho et al. (2004) (2005xxxvi) Proteobacteria Garrity et al. 2005 – Valid publication: Validation Burkholderiales Garrity et al. 2006, 1VP – Valid publication: Vali- List no. 106 – Effective publication: Garrity et al. (2005i) dation List no. 107 – Effective publication: Garrity et al. (2005xxiii) New classes Caldilineales Yamada et al. 2006, 1339VP VP Alphaproteobacteria Garrity et al. 2006, 1 – Valid publication: Campylobacterales Garrity et al. 2006, 1VP – Valid publication: Validation List no. 107 – Effective publication: Garrity et al. Validation List no. 107 – Effective publication: Garrity et al. (2005xv) (2005xxxixi) VP Anaerolineae Yamada et al. 2006, 1336 Cardiobacteriales Garrity et al. 2005, 2235VP – Valid publica- Betaproteobacteria Garrity et al. 2006, 1VP – Valid publication: tion: Validation List no. 106 – Effective publication: Garrity Validation List no. 107 – Effective publication: Garrity et al. -

Genome Sequence and Description of Mannheimia Massilioguelmaensis Sp
Genome sequence and description of Mannheimia massilioguelmaensis sp. nov. L. Hadjadj, Ahmed Aimen Bentorki, C. Michelle, K. Amoura, Abdelghani Djahoudi, Jean-Marc Rolain To cite this version: L. Hadjadj, Ahmed Aimen Bentorki, C. Michelle, K. Amoura, Abdelghani Djahoudi, et al.. Genome sequence and description of Mannheimia massilioguelmaensis sp. nov.. New Microbes and New Infec- tions, Wiley Online Library 2015, 8, pp.131-136. 10.1016/j.nmni.2015.10.005. hal-01772904 HAL Id: hal-01772904 https://hal-amu.archives-ouvertes.fr/hal-01772904 Submitted on 20 Apr 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - NoDerivatives| 4.0 International License TAXONOGENOMICS: GENOME OF A NEW ORGANISM Genome sequence and description of Mannheimia massilioguelmaensis sp. nov. L. Hadjadj1, A. A. Bentorki2, C. Michelle1, K. Amoura3, A. Djahoudi3 and J.-M. Rolain1 1) Unité de recherche sur les maladies infectieuses et tropicales émergentes (URMITE), UMR CNRS, IHU Méditerranée Infection, Faculté de Médecine et de Pharmacie, Aix-Marseille-Université, Marseille, France, 2) Laboratoire de Microbiologie, CHU Dorban and 3) Laboratoire de Microbiologie, Faculté de Médecine, Université Badji Mokhtar, Annaba, Algeria Abstract Strain MG13T sp. -
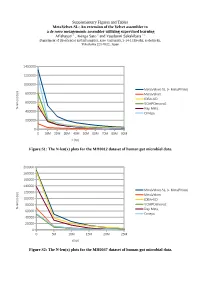
Supplementary Figures and Tables Metavelvet-SL
Supplementary Figures and Tables MetaVelvet-SL: An extension of the Velvet assembler to a de novo metagenomic assembler utilizing supervised learning Afiahayati 1 , Kengo Sato 1 and Yasubumi Sakakibara 1∗ Department of Biosciences and Informatics, Keio University, 3-14-1 Hiyoshi, Kohoku-ku, Yokohama 223-8522, Japan 1400000 1200000 1000000 MetaVelvet-SL (+ MetaPhlAn) ) p 800000 b MetaVelvet ( ) x ( IDBA-UD n 600000 e SOAPDenovo2 l - N Ray Meta 400000 Omega 200000 0 0 10M 20M 30M 40M 50M 60M 70M 80M 90M x (bp) Figure S1: The N-len(x) plots for the MH0012 dataset of human gut microbial data. 200000 180000 160000 140000 MetaVelvet-SL (+ MetaPhlAn) ) 120000 p b MetaVelvet ( ) 100000 x IDBA-UD ( n e l 80000 SOAPDenovo2 - N Ray Meta 60000 Omega 40000 20000 0 0 5M 10M 15M 20M 25M x(bp) Figure S2: The N-len(x) plots for the MH0047 dataset of human gut microbial data. 600000 500000 400000 MetaVelvet-SL (+ MetaPhlAn) ) p b MetaVelvet ( ) 300000 x IDBA-UD ( n e l SOAPDenovo2 - N 200000 Ray Meta Omega 100000 0 0 10M 20M 30M 40M 50M 60M 70M 80M 90M x (bp) Figure S3: The N-len(x) plots for the SRS017227 dataset of human gut microbial data. 800000 700000 600000 500000 MetaVelvet-SL (+ MetaPhlAn) ) p b MetaVelvet ( ) 400000 x IDBA-UD ( n e l SOAPDenovo2 - 300000 N Ray Meta 200000 Omega 100000 0 0 5M 10M 15M 20M 25M 30M 35M 40M 45M x (bp) Figure S4: The N-len(x) plots for the SRS018661 dataset of human gut microbial data. Formula to identify unique nodes in Velvet (Zerbino and Birney, 2008) ̄x2 ρ2− log2 2 F ( ̄x ,n,ρ )= +n 2 2 where ̄x = the coverage of node ρ = expected coverage of subgraph n = the length of node A node is “unique”, if its F > 5 . -

Reorganising the Order Bacillales Through Phylogenomics
Systematic and Applied Microbiology 42 (2019) 178–189 Contents lists available at ScienceDirect Systematic and Applied Microbiology jou rnal homepage: http://www.elsevier.com/locate/syapm Reorganising the order Bacillales through phylogenomics a,∗ b c Pieter De Maayer , Habibu Aliyu , Don A. Cowan a School of Molecular & Cell Biology, Faculty of Science, University of the Witwatersrand, South Africa b Technical Biology, Institute of Process Engineering in Life Sciences, Karlsruhe Institute of Technology, Germany c Centre for Microbial Ecology and Genomics, University of Pretoria, South Africa a r t i c l e i n f o a b s t r a c t Article history: Bacterial classification at higher taxonomic ranks such as the order and family levels is currently reliant Received 7 August 2018 on phylogenetic analysis of 16S rRNA and the presence of shared phenotypic characteristics. However, Received in revised form these may not be reflective of the true genotypic and phenotypic relationships of taxa. This is evident in 21 September 2018 the order Bacillales, members of which are defined as aerobic, spore-forming and rod-shaped bacteria. Accepted 18 October 2018 However, some taxa are anaerobic, asporogenic and coccoid. 16S rRNA gene phylogeny is also unable to elucidate the taxonomic positions of several families incertae sedis within this order. Whole genome- Keywords: based phylogenetic approaches may provide a more accurate means to resolve higher taxonomic levels. A Bacillales Lactobacillales suite of phylogenomic approaches were applied to re-evaluate the taxonomy of 80 representative taxa of Bacillaceae eight families (and six family incertae sedis taxa) within the order Bacillales. -

(ST). the Table
1 SUPPLEMENTAL MATERIALS Growth Media Modern Condition Seawater Freshwater Light T°C Atmosphere AMCONA medium BG11 medium – Synechococcus – – Synechocystis – [C] in PAL [C] in the [C] in the Gas Nutrients Modern Nutrients (ppm) Medium Medium Ocean NaNO3 CO2 ~407.8 Na2SO4 25.0mM 29mM Nitrogen 50 Standard (17.65mM) μmol (ST) NaNO NaNO 20°C O ~209’460 Nitrogen 3 3 MgSO 0.304mM photon 2 (549µM) (13.7µM) 4 /m2s FeCl3 6.56µM 2nM Ammonium 0.6g/L ZnSO 254nM 0.5nM ferric stock N ~780’790 4 2 citrate (10ml NaMoO 105nM 105nM 4 green stock/1L) 2 Table S 1 Description of the experimental condition defined as Standard Condition (ST). The table 3 shows the concentrations of fundamental elements, such as C, N, S, and Fe used for the AMCONA 4 seawater medium (Fanesi et al., 2014) and BG11 freshwater medium (Stanier et al., 1971) flushing air 5 with using air pump (KEDSUM-310 8W pump; Xiolan, China) Growth Media Possible Proterozoic T° Condition Modified Seawater Modified Freshwater Light Atmosphere AMCONA medium BG11 medium C – Synechococcus – – Synechocystis – [C] in [C] in PPr Nutrients PPr Nutrients Medium Gas ppm Medium NH Cl Na SO 3mM Nitrogen 4 3 2 4 (0.0035mM) CO 2 10’000ppm (20%) NH Cl Possible (~ 2’450% Nitrogen 4 3 MgSO 0.035mM 50 with 20ml/ (100µM) 4 Proterozoic PAL) μmol 20° min (PPr) photon C O2 20’000ppm /m2s (in Air) (~ 10% FeCl 200nM with 5ml/ 3 PAL) Ammonium 0.6g/L stock min ferric 10ml N ZnSO 0.0nM 2 4 citrate green stock/1L (100%) Base gas with NaMoO4 10.5nM 200ml/min 6 Table S 2 Description of the experimental condition defined as Possible Proterozoic Condition (PPr). -

Noncontiguous Finished Genome Sequence and Description Of
TAXONOGENOMICS: GENOME OF A NEW ORGANISM Noncontiguous finished genome sequence and description of Bacillus andreraoultii strain SIT1T sp. nov. S. I. Traore1,2, T. Cimmino1, J.-C. Lagier1, S. Khelaifia1, S. Brah3, C. Michelle1, A. Caputo1, B. A. Diallo4, P.-E. Fournier1, D. Raoult1,5 and J. M. Rolain1 1) Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes, UM 63, CNRS 7278, IRD 198, Inserm U1095, Institut Hospitalo-Universitaire Méditerranée-Infection, Faculté de médecine, Aix-Marseille Université, Marseille, France, 2) Département d’Epidémiologie des Affections Parasitaires, Faculté de médecine et de pharmacie de Bamako, Mali, 3) Hôpital National de Niamey, 4) Laboratoire de Microbiologie, Département de Biologie, Université Abdou Moumouni de Niamey, Niamey, Niger and 5) Special Infectious Agents Unit, King Fahd Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia Abstract Bacillus andreraoultii strain SIT1T (= CSUR P1162 = DSM 29078) is the type strain of B. andreraoultii sp. nov. This bacterium was isolated from the stool of a 2-year-old Nigerian boy with a severe form of kwashiorkor. Bacillus andreraoultii is an aerobic, Gram-positive rod. We describe here the features of this bacterium, together with the complete genome sequencing and annotation. The 4 092 130 bp long genome contains 3718 protein-coding and 116 RNA genes. New Microbes and New Infections © 2015 The Authors. Published by Elsevier Ltd on behalf of European Society of Clinical Microbiology and Infectious Diseases. Keywords: Culturomics, genome Original Submission: 14 September 2015; Revised Submission: 15 December 2015; Accepted: 16 December 2015 Article published online: 23 December 2015 gene-based phylogeny [5], G+C content and DNA-DNA hy- Corresponding author: J. -

T H È S E Rita Abou Abdallah
81,9(56,7e D’AIX0$56(,//( )$&8/7e'(0e'(&,1('(0$56(,//( 7+Ê6( 6RXWHQXHSXEOLTXHPHQWOH-XLOOHW en vue de l’obtention du titre de Docteur de l’Université d’Aix0DUVHLOOH 6SpFLDOLWp0DODGLHV,QIHFWLHXVHV SDU 5LWD$ERX$EGDOODK /D*pQRPLTXH 8Q2XWLO5REXVWHHW(PHUJHQW8WLOLVp GDQVOD7D[RQRPLH%DFWpULHQQHHW O’analyse Comparative &RPSRVLWLRQGXMXU\ 3URIHVVHXU%/$6&2/$ 8QLYHUVLWp$L[0DUVHLOOH 3UpVLGHQWGX-XU\ 3URIHVVHXU00$85,1 8QLYHUVLWpGH*UHQREOH 5DSSRUWHXU 3URIHVVHXU35(1(672 8QLYHUVLWpGH*UHQREOH 5DSSRUWHXU 3URIHVVHXU3()2851,(5 Université d’Aix0DUVHLOOH 'LUHFWHXUGH7KqVH 8059,7520( $L[0DUVHLOOH8QLYHUVLWp,QVWLWXWGH5HFKHUFKHSRXUOH'pYHORSSHPHQW $VVLVWDQFH3XEOLTXH–+{SLWDX[GH0DUVHLOOH $3+0 6HUYLFHGH6DQWp GHV$UPpHV,QVWLWXW+RVSLWDOR8QLYHUVLWDLUH0pGLWHUUDQpH,QIHFWLRQ 5HPHUFLHPHQWV Je tiens à remercier Professeur Pierre-Edourad Fournier, pour son encadrement et enseignement au cours de ces dernières années. Ses conseils et encouragements ont été des éléments moteurs pour moi et déterminants pour le déroulement de cette thèse. Votre calme, respect, humanité et gentillesse envers vos étudiants sont gravés dans mon cœur. Finalement, je vous remercie pour votre support et confiance tout au long de ces trois années. Vous aurez toujours une place dans mon esprit et mon cœur. Je remercie Professeur Didier Raoult pour m’avoir accueillie au sein de l’Institut Hospitalo-Universitaire, Méditerranée-Infection. C’est dans un environnement stimulant intellectuellement que mon goût pour la recherche est né. Je remercie Professeur Bernard La Scola, d’avoir accepté de présider au sein du jury de cette Thèse. Je remercie Professeur Max Maurin et Patricia Renesto d’avoir accepté de siéger au sein du jury de cette Thèse et pour l’intérêt porté à ce travail. Je remercie ma mère et ma sœur, pour leurs supports et conseils quotidiens.