Phylogenetics of Seed Plants: an Analysis of Nucleotide Sequences from the Plastid Gene Rbcl James F
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

I Scope and Importance of Taxonomy. Classification of Angiosperms- Bentham and Hooker System & Cronquist
Syllabus: 2020-2021 Unit – I Scope and importance of Taxonomy. Classification of Angiosperms- Bentham and Hooker system & Cronquist. Flora, revision and Monographs. Botanical nomenclature (ICBN), Taxonomic hierarchy, typification, principles of priority, publication, Keys and their types, Preparation and role of Herbarium. Importance of Botanical gardens. PLANT KINGDOM Amongst plants nearly 15,000 species belong to Mosses and Liverworts, 12,700 Ferns and their allies, 1,079 Gymnosperms and 295,383 Angiosperms (belonging to about 485 families and 13,372 genera), considered to be the most recent and vigorous group of plants that have occurred on earth. Angiosperms occupy the majority of the terrestrial space on earth, and are the major components of the world‘s vegetation. Brazil (First) and Colombia (second), both located in the tropics considered to be countries with the most diverse angiosperms floras China (Third) even though the main part of her land is not located in the tropics, the number of angiosperms still occupies the third place in the world. In INDIA there are about 18042 species of flowering plants approximately 320 families, 40 genera and 30,000 species. IUCN Red list Categories: EX –Extinct; EW- Extinct in the Wild-Threatened; CR -Critically Endangered; VU- Vulnerable Angiosperm (Flowering Plants) SPECIES RICHNESS AROUND THE WORLD PLANT CLASSIFICATION Historia Plantarum - the earliest surviving treatise on plants in which Theophrastus listed the names of over 500 plant species. Artificial system of Classification Theophrastus attempted common groupings of folklore combined with growth form such as ( Tree Shrub; Undershrub); or Herb. Or (Annual and Biennials plants) or (Cyme and Raceme inflorescences) or (Archichlamydeae and Meta chlamydeae) or (Upper or Lower ovarian ). -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

Arthur Monrad Johnson Colletion of Botanical Drawings
http://oac.cdlib.org/findaid/ark:/13030/kt7489r5rb No online items Arthur Monrad Johnson colletion of botanical drawings 1914-1941 Processed by Pat L. Walter. Louise M. Darling Biomedical Library History and Special Collections Division History and Special Collections Division UCLA 12-077 Center for Health Sciences Box 951798 Los Angeles, CA 90095-1798 Phone: 310/825-6940 Fax: 310/825-0465 Email: [email protected] URL: http://www.library.ucla.edu/libraries/biomed/his/ ©2008 The Regents of the University of California. All rights reserved. Arthur Monrad Johnson colletion 48 1 of botanical drawings 1914-1941 Descriptive Summary Title: Arthur Monrad Johnson colletion of botanical drawings, Date (inclusive): 1914-1941 Collection number: 48 Creator: Johnson, Arthur Monrad 1878-1943 Extent: 3 boxes (2.5 linear feet) Repository: University of California, Los Angeles. Library. Louise M. Darling Biomedical Library History and Special Collections Division Los Angeles, California 90095-1490 Abstract: Approximately 1000 botanical drawings, most in pen and black ink on paper, of the structural parts of angiosperms and some gymnosperms, by Arthur Monrad Johnson. Many of the illustrations have been published in the author's scientific publications, such as his "Taxonomy of the Flowering Plants" and articles on the genus Saxifraga. Dr. Johnson was both a respected botanist and an accomplished artist beyond his botanical subjects. Physical location: Collection stored off-site (Southern Regional Library Facility): Advance notice required for access. Language of Material: Collection materials in English Preferred Citation [Identification of item], Arthur Monrad Johnson colletion of botanical drawings (Manuscript collection 48). Louise M. Darling Biomedical Library History and Special Collections Division, University of California, Los Angeles. -

News Quarterly
Heather News Quarterly Volume 34 Number 4 Issue #136 Fall 2011 North American Heather Society Your guess is as good as mine Donald Mackay........................1 Editing the heather garden Ella May T. Wulff............................6 In memoriam: Judith Wiksten Ella May T. Wulff....................18 My other favorite heathers Irene Henson...............................19 Erica carnea ‘Golden Starlet’ Pat Hoffman...............................24 Misleading advertising department...........................................27 Calendar........................................................................................28 Index 2011.....................................................................................28 North American Heather Society Membership Chair Ella May Wulff, Knolls Drive 2299 Wooded Philomath, OR 97370-5908 RETURN SERVICE REQUESTED issn 1041-6838 Heather News Quarterly, all rights reserved, is published quarterly by the North American Heather Society, a tax exempt organization. The purpose of The Society The Information Page is the: (1) advancement and study of the botanical genera Calluna, Cassiope, Daboecia, Erica, and Phyllodoce, commonly called heather, and related genera; (2) HOW TO GET THE Latest heather INFORMation dissemination of information on heather; and (3) promotion of fellowship among BROWSE NAHS website – www.northamericanheathersociety.org those interested in heather. READ Heather News Quarterly by NAHS CHS NEWS by CHS Heather Clippings by HERE NAHS Board of Directors (2010-2011) Heather Drift by VIHS Heather News by MCHS Heather Notes by NEHS Heather & Yon by OHS PRESIDENT ATTEND Society and Chapter meetings (See The Calendar on page 28) Karla Lortz, 502 E Haskell Hill Rd., Shelton, WA 98584-8429, USA 360-427-5318, [email protected] HOW TO GET PUBLISHED IN HEATHER NEWS QUARTERLY FIRST VICE-PRESIDENT CONTACT Stefani McRae-Dickey, Editor of Heather News Quarterly Don Jewett, 2655 Virginia Ct., Fortuna, CA 95540, USA [email protected] 541-929-7988. -

Phylogeny of Malpighiaceae: Evidence from Chloroplast NDHF and TRNL-F Nucleotide Sequences
Phylogeny of Malpighiaceae: Evidence from Chloroplast NDHF and TRNL-F Nucleotide Sequences The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Davis, Charles C., William R. Anderson, and Michael J. Donoghue. 2001. Phylogeny of Malpighiaceae: Evidence from chloroplast NDHF and TRNL-F nucleotide sequences. American Journal of Botany 88(10): 1830-1846. Published Version http://dx.doi.org/10.2307/3558360 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:2674790 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA American Journal of Botany 88(10): 1830±1846. 2001. PHYLOGENY OF MALPIGHIACEAE: EVIDENCE FROM CHLOROPLAST NDHF AND TRNL-F NUCLEOTIDE SEQUENCES1 CHARLES C. DAVIS,2,5 WILLIAM R. ANDERSON,3 AND MICHAEL J. DONOGHUE4 2Department of Organismic and Evolutionary Biology, Harvard University Herbaria, 22 Divinity Avenue, Cambridge, Massachusetts 02138 USA; 3University of Michigan Herbarium, North University Building, Ann Arbor, Michigan 48109-1057 USA; and 4Department of Ecology and Evolutionary Biology, Yale University, P.O. Box 208106, New Haven, Connecticut 06520 USA The Malpighiaceae are a family of ;1250 species of predominantly New World tropical ¯owering plants. Infrafamilial classi®cation has long been based on fruit characters. Phylogenetic analyses of chloroplast DNA nucleotide sequences were analyzed to help resolve the phylogeny of Malpighiaceae. A total of 79 species, representing 58 of the 65 currently recognized genera, were studied. -

(Asteraceae): a Relict Genus of Cichorieae?
Anales del Jardín Botánico de Madrid Vol. 65(2): 367-381 julio-diciembre 2008 ISSN: 0211-1322 Warionia (Asteraceae): a relict genus of Cichorieae? by Liliana Katinas1, María Cristina Tellería2, Alfonso Susanna3 & Santiago Ortiz4 1 División Plantas Vasculares, Museo de La Plata, Paseo del Bosque s/n, 1900 La Plata, Argentina. [email protected] 2 Laboratorio de Sistemática y Biología Evolutiva, Museo de La Plata, Paseo del Bosque s/n, 1900 La Plata, Argentina. [email protected] 3 Instituto Botánico de Barcelona, Pg. del Migdia s.n., 08038 Barcelona, Spain. [email protected] 4 Laboratorio de Botánica, Facultade de Farmacia, Universidade de Santiago, 15782 Santiago de Compostela, Spain. [email protected] Abstract Resumen Katinas, L., Tellería, M.C., Susanna, A. & Ortiz, S. 2008. Warionia Katinas, L., Tellería, M.C., Susanna, A. & Ortiz, S. 2008. Warionia (Asteraceae): a relict genus of Cichorieae? Anales Jard. Bot. Ma- (Asteraceae): un género relicto de Cichorieae? Anales Jard. Bot. drid 65(2): 367-381. Madrid 65(2): 367-381 (en inglés). The genus Warionia, with its only species W. saharae, is endemic to El género Warionia, y su única especie, W. saharae, es endémico the northwestern edge of the African Sahara desert. This is a some- del noroeste del desierto africano del Sahara. Es una planta seme- what thistle-like aromatic plant, with white latex, and fleshy, pin- jante a un cardo, aromática, con látex blanco y hojas carnosas, nately-partite leaves. Warionia is in many respects so different from pinnatipartidas. Warionia es tan diferente de otros géneros de any other genus of Asteraceae, that it has been tentatively placed Asteraceae que fue ubicada en las tribus Cardueae, Cichorieae, in the tribes Cardueae, Cichorieae, Gundelieae, and Mutisieae. -
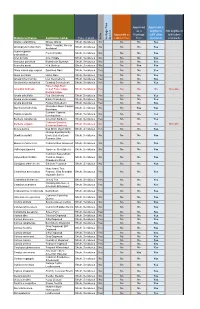
Botanical Name Common Name
Approved Approved & as a eligible to Not eligible to Approved as Frontage fulfill other fulfill other Type of plant a Street Tree Tree standards standards Heritage Tree Tree Heritage Species Botanical Name Common name Native Abelia x grandiflora Glossy Abelia Shrub, Deciduous No No No Yes White Forsytha; Korean Abeliophyllum distichum Shrub, Deciduous No No No Yes Abelialeaf Acanthropanax Fiveleaf Aralia Shrub, Deciduous No No No Yes sieboldianus Acer ginnala Amur Maple Shrub, Deciduous No No No Yes Aesculus parviflora Bottlebrush Buckeye Shrub, Deciduous No No No Yes Aesculus pavia Red Buckeye Shrub, Deciduous No No Yes Yes Alnus incana ssp. rugosa Speckled Alder Shrub, Deciduous Yes No No Yes Alnus serrulata Hazel Alder Shrub, Deciduous Yes No No Yes Amelanchier humilis Low Serviceberry Shrub, Deciduous Yes No No Yes Amelanchier stolonifera Running Serviceberry Shrub, Deciduous Yes No No Yes False Indigo Bush; Amorpha fruticosa Desert False Indigo; Shrub, Deciduous Yes No No No Not eligible Bastard Indigo Aronia arbutifolia Red Chokeberry Shrub, Deciduous Yes No No Yes Aronia melanocarpa Black Chokeberry Shrub, Deciduous Yes No No Yes Aronia prunifolia Purple Chokeberry Shrub, Deciduous Yes No No Yes Groundsel-Bush; Eastern Baccharis halimifolia Shrub, Deciduous No No Yes Yes Baccharis Summer Cypress; Bassia scoparia Shrub, Deciduous No No No Yes Burning-Bush Berberis canadensis American Barberry Shrub, Deciduous Yes No No Yes Common Barberry; Berberis vulgaris Shrub, Deciduous No No No No Not eligible European Barberry Betula pumila -

Biogeography and Diversification of Brassicales
Molecular Phylogenetics and Evolution 99 (2016) 204–224 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Biogeography and diversification of Brassicales: A 103 million year tale ⇑ Warren M. Cardinal-McTeague a,1, Kenneth J. Sytsma b, Jocelyn C. Hall a, a Department of Biological Sciences, University of Alberta, Edmonton, Alberta T6G 2E9, Canada b Department of Botany, University of Wisconsin, Madison, WI 53706, USA article info abstract Article history: Brassicales is a diverse order perhaps most famous because it houses Brassicaceae and, its premier mem- Received 22 July 2015 ber, Arabidopsis thaliana. This widely distributed and species-rich lineage has been overlooked as a Revised 24 February 2016 promising system to investigate patterns of disjunct distributions and diversification rates. We analyzed Accepted 25 February 2016 plastid and mitochondrial sequence data from five gene regions (>8000 bp) across 151 taxa to: (1) Available online 15 March 2016 produce a chronogram for major lineages in Brassicales, including Brassicaceae and Arabidopsis, based on greater taxon sampling across the order and previously overlooked fossil evidence, (2) examine Keywords: biogeographical ancestral range estimations and disjunct distributions in BioGeoBEARS, and (3) determine Arabidopsis thaliana where shifts in species diversification occur using BAMM. The evolution and radiation of the Brassicales BAMM BEAST began 103 Mya and was linked to a series of inter-continental vicariant, long-distance dispersal, and land BioGeoBEARS bridge migration events. North America appears to be a significant area for early stem lineages in the Brassicaceae order. Shifts to Australia then African are evident at nodes near the core Brassicales, which diverged Cleomaceae 68.5 Mya (HPD = 75.6–62.0). -

Number 3, Spring 1998 Director’S Letter
Planning and planting for a better world Friends of the JC Raulston Arboretum Newsletter Number 3, Spring 1998 Director’s Letter Spring greetings from the JC Raulston Arboretum! This garden- ing season is in full swing, and the Arboretum is the place to be. Emergence is the word! Flowers and foliage are emerging every- where. We had a magnificent late winter and early spring. The Cornus mas ‘Spring Glow’ located in the paradise garden was exquisite this year. The bright yellow flowers are bright and persistent, and the Students from a Wake Tech Community College Photography Class find exfoliating bark and attractive habit plenty to photograph on a February day in the Arboretum. make it a winner. It’s no wonder that JC was so excited about this done soon. Make sure you check of themselves than is expected to seedling selection from the field out many of the special gardens in keep things moving forward. I, for nursery. We are looking to propa- the Arboretum. Our volunteer one, am thankful for each and every gate numerous plants this spring in curators are busy planting and one of them. hopes of getting it into the trade. preparing those gardens for The magnolias were looking another season. Many thanks to all Lastly, when you visit the garden I fantastic until we had three days in our volunteers who work so very would challenge you to find the a row of temperatures in the low hard in the garden. It shows! Euscaphis japonicus. We had a twenties. There was plenty of Another reminder — from April to beautiful seven-foot specimen tree damage to open flowers, but the October, on Sunday’s at 2:00 p.m. -

The in Vitro Evaluation of Antibacterial and Antifungal Activities of Ripe Fruit Extracts (Pericarp and Seed) of Bunchosia Armeniaca (Cav.) DC
ASIAN JOURNAL OF PHARMACOGNOSY Research Article Asian J. Pharmacogn 3(3):20-28 © 2019, Asian Society of Pharmacognosy. All Rights Reserved. eISSN-0128-1119 The in vitro evaluation of antibacterial and antifungal activities of ripe fruit extracts (pericarp and seed) of Bunchosia armeniaca (Cav.) DC. Mackingsley Kushan Dassanayake University of Wolverhampton, Wulfruna Street, Wolverhampton, WV1 1LY, UK *For correspondence: [email protected] _________________________________ Abstract: Plants are rich sources of phytoconstituents that have the ability to contribute biological activities. Indigenously prepared crude extracts of medicinal plants have been used as medications to treat infections caused by pathogenic microorganisms by people of various ethnic origins for many centuries in folkloric medicine. The research in discovering new and innovative antimicrobial compounds from plants has received increased concern due to the global rise of antimicrobial resistance. The present study aims to evaluate the in vitro antimicrobial potential of pericarp and seed extracts of ripe fruits associated with a native South American plant specimen known as Bunchosia armeniaca. The antibacterial and antifungal activities of aqueous and ethanolic crude extracts with concentrations ranging from 1000 to 125 µg/ml were determined against bacterial pathogens of Staphylococcus aureus, Escherichia coli, Bacillus subtilis, Pseudomonas aeruginosa, Klebsiella pneumoniae, Streptococcus pyogenes, viridans group streptococci, and fungal pathogens Candida albicans and Aspergillus fumigates by performing the agar well diffusion assay. The pericarp extract did not exhibit any antimicrobial activity while the aqueous seed extract was only active against Escherichia coli, which indicated a mean inhibition zone diameter of 16.2 mm at its highest concentration. The minimum inhibitory concentration of the seed extract was 250 µg/ml. -

Chapter 6 ENUMERATION
Chapter 6 ENUMERATION . ENUMERATION The spermatophytic plants with their accepted names as per The Plant List [http://www.theplantlist.org/ ], through proper taxonomic treatments of recorded species and infra-specific taxa, collected from Gorumara National Park has been arranged in compliance with the presently accepted APG-III (Chase & Reveal, 2009) system of classification. Further, for better convenience the presentation of each species in the enumeration the genera and species under the families are arranged in alphabetical order. In case of Gymnosperms, four families with their genera and species also arranged in alphabetical order. The following sequence of enumeration is taken into consideration while enumerating each identified plants. (a) Accepted name, (b) Basionym if any, (c) Synonyms if any, (d) Homonym if any, (e) Vernacular name if any, (f) Description, (g) Flowering and fruiting periods, (h) Specimen cited, (i) Local distribution, and (j) General distribution. Each individual taxon is being treated here with the protologue at first along with the author citation and then referring the available important references for overall and/or adjacent floras and taxonomic treatments. Mentioned below is the list of important books, selected scientific journals, papers, newsletters and periodicals those have been referred during the citation of references. Chronicles of literature of reference: Names of the important books referred: Beng. Pl. : Bengal Plants En. Fl .Pl. Nepal : An Enumeration of the Flowering Plants of Nepal Fasc.Fl.India : Fascicles of Flora of India Fl.Brit.India : The Flora of British India Fl.Bhutan : Flora of Bhutan Fl.E.Him. : Flora of Eastern Himalaya Fl.India : Flora of India Fl Indi. -

First Steps Towards a Floral Structural Characterization of the Major Rosid Subclades
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2006 First steps towards a floral structural characterization of the major rosid subclades Endress, P K ; Matthews, M L Abstract: A survey of our own comparative studies on several larger clades of rosids and over 1400 original publications on rosid flowers shows that floral structural features support to various degrees the supraordinal relationships in rosids proposed by molecular phylogenetic studies. However, as many apparent relationships are not yet well resolved, the structural support also remains tentative. Some of the features that turned out to be of interest in the present study had not previously been considered in earlier supraordinal studies. The strongest floral structural support is for malvids (Brassicales, Malvales, Sapindales), which reflects the strong support of phylogenetic analyses. Somewhat less structurally supported are the COM (Celastrales, Oxalidales, Malpighiales) and the nitrogen-fixing (Cucurbitales, Fagales, Fabales, Rosales) clades of fabids, which are both also only weakly supported in phylogenetic analyses. The sister pairs, Cucurbitales plus Fagales, and Malvales plus Sapindales, are structurally only weakly supported, and for the entire fabids there is no clear support by the present floral structural data. However, an additional grouping, the COM clade plus malvids, shares some interesting features but does not appear as a clade in phylogenetic analyses. Thus it appears that the deepest split within eurosids- that between fabids and malvids - in molecular phylogenetic analyses (however weakly supported) is not matched by the present structural data. Features of ovules including thickness of integuments, thickness of nucellus, and degree of ovular curvature, appear to be especially interesting for higher level relationships and should be further explored.