Binding Interaction Analysis of Neuromedin U Receptor 1 with the Native Protein Neuromedin U
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ILC2 Activation by Protozoan Commensal Microbes
International Journal of Molecular Sciences Review ILC2 Activation by Protozoan Commensal Microbes Kyle Burrows 1 , Louis Ngai 1 , Flora Wong 1,2, David Won 1 and Arthur Mortha 1,* 1 University of Toronto, Department of Immunology, Toronto, ON M5S 1A8, Canada; [email protected] (K.B.) [email protected] (L.N.); fl[email protected] (F.W.); [email protected] (D.W.) 2 Ranomics, Inc. Toronto, ON M5G 1X5, Canada * Correspondence: [email protected] Received: 3 September 2019; Accepted: 27 September 2019; Published: 30 September 2019 Abstract: Group 2 innate lymphoid cells (ILC2s) are a member of the ILC family and are involved in protective and pathogenic type 2 responses. Recent research has highlighted their involvement in modulating tissue and immune homeostasis during health and disease and has uncovered critical signaling circuits. While interactions of ILC2s with the bacterial microbiome are rather sparse, other microbial members of our microbiome, including helminths and protozoans, reveal new and exciting mechanisms of tissue regulation by ILC2s. Here we summarize the current field on ILC2 activation by the tissue and immune environment and highlight particularly new intriguing pathways of ILC2 regulation by protozoan commensals in the intestinal tract. Keywords: ILC2; protozoa; Trichomonas; Tritrichomonas musculis; mucosal immunity; taste receptors; succinate; intestinal immunity; type 2 immunity; commensals 1. The ILC Lineage 1.1. The Family of Innate Lymphoid Cells Research over the last decade has redirected focus away from classical immune cell interactions within lymphoid tissues towards immunity within non-lymphoid tissues. Within these tissues, immune interactions involve local adaptation and rapid responses by tissue-resident immune cells. -

Neural Regulation of Interactions Between Group 2 Innate Lymphoid Cells and Pulmonary Immune Cells
Prime Archives in Immunology: 2nd Edition Book Chapter Neural Regulation of Interactions between Group 2 Innate Lymphoid Cells and Pulmonary Immune Cells Weiwei Chen1,2, Qiang Shu2 and Jie Fan1,3,4* 1Department of Surgery, University of Pittsburgh School of Medicine, USA 2The Children‟s Hospital, Zhejiang University School of Medicine, National Clinical Research Center for Child Health, China 3Research and Development, Veterans Affairs Pittsburgh Healthcare System, USA 4McGowan Institute for Regenerative Medicine, University of Pittsburgh, USA *Corresponding Author: Jie Fan, Department of Surgery, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA Published January 18, 2021 This Book Chapter is a republication of an article published by Jie Fan, et al. at Frontiers in Immunology in October 2020. (Chen W, Shu Q and Fan J (2020) Neural Regulation of Interactions Between Group 2 Innate Lymphoid Cells and Pulmonary Immune Cells. Front. Immunol. 11:576929. doi: 10.3389/fimmu.2020.576929) How to cite this book chapter: Weiwei Chen, Qiang Shu, Jie Fan. Neural Regulation of Interactions between Group 2 Innate Lymphoid Cells and Pulmonary Immune Cells. In: Ajmal Khan, Ahmed Al-Harrasi, editors. Prime Archives in Immunology: 2nd Edition. Hyderabad, India: Vide Leaf. 2021. 1 www.videleaf.com Prime Archives in Immunology: 2nd Edition © The Author(s) 2021. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License(http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Author Contributions: WC collected the data and drafted the manuscript. WC, QS, and JF conceived and designed the study. -

Guthrie Cdna Resource Center
cDNA Resource Center cDNA Resource Center Catalog cDNA Resource Center Missouri University of Science and Technology 400 W 11th Rolla, MO 65409 TEL: (573) 341-7610 FAX: (573) 341-7609 EMAIL: [email protected] www.cdna.org September, 2008 1 cDNA Resource Center Visit our web site for product updates 2 cDNA Resource Center The cDNA Resource Center The cDNA Resource Center is a service provided by the faculty of the Department of Biological Sciences of Missouri University of Science and Technology. The purpose of the cDNA Resource Center is to further scientific investigation by providing cDNA clones of human proteins involved in signal transduction processes. This is achieved by providing high quality clones for important signaling proteins in a timely manner. By high quality, we mean that the clones are • Sequence verified • Propagated in a versatile vector useful in bacterial and mammalian systems • Free of extraneous 3' and 5' untranslated regions • Expression verified (in most cases) by coupled in vitro transcription/translation assays • Available in wild-type, epitope-tagged and common mutant forms (e.g., constitutively- active or dominant negative) By timely, we mean that the clones are • Usually shipped within a day from when you place your order. Clones can be ordered from our web pages, by FAX or by phone. Within the United States, clones are shipped by overnight courier (FedEx); international orders are shipped International Priority (FedEx). The clones are supplied for research purposes only. Details on use of the material are included on the Material Transfer Agreement (page 3). Clones are distributed by agreement in Invitrogen's pcDNA3.1+ vector. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors
Ecology and Evolutionary Biology 2021; 6(3): 53-77 http://www.sciencepublishinggroup.com/j/eeb doi: 10.11648/j.eeb.20210603.11 ISSN: 2575-3789 (Print); ISSN: 2575-3762 (Online) An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors Miguel Angel Fuertes*, Carlos Alonso Department of Microbiology, Centre for Molecular Biology “Severo Ochoa”, Spanish National Research Council and Autonomous University, Madrid, Spain Email address: *Corresponding author To cite this article: Miguel Angel Fuertes, Carlos Alonso. An Evolutionary Based Strategy for Predicting Rational Mutations in G Protein-Coupled Receptors. Ecology and Evolutionary Biology. Vol. 6, No. 3, 2021, pp. 53-77. doi: 10.11648/j.eeb.20210603.11 Received: April 24, 2021; Accepted: May 11, 2021; Published: July 13, 2021 Abstract: Capturing conserved patterns in genes and proteins is important for inferring phenotype prediction and evolutionary analysis. The study is focused on the conserved patterns of the G protein-coupled receptors, an important superfamily of receptors. Olfactory receptors represent more than 2% of our genome and constitute the largest family of G protein-coupled receptors, a key class of drug targets. As no crystallographic structures are available, mechanistic studies rely on the use of molecular dynamic modelling combined with site-directed mutagenesis data. In this paper, we hypothesized that human-mouse orthologs coding for G protein-coupled receptors maintain, at speciation events, shared compositional structures independent, to some extent, of their percent identity as reveals a method based in the categorization of nucleotide triplets by their gross composition. The data support the consistency of the hypothesis, showing in ortholog G protein-coupled receptors the presence of emergent shared compositional structures preserved at speciation events. -
* Supplementary Table 3B. Complete List of Lymphocytic-Associated Genes
Supplementary Table 3b. Complete list of lymphocytic-associated genes for the HER2+I subtype based on the SNR score (p-val <= 0.005 and FDR<=0.05). A positive score represents genes up-regulated in HER2+I and a negative score represents genes up-regulated in HER2+NI. The first 9 genes, marked with *, are chemokines near the HER2+ amplicon at chr17q12. The remaining are sorted based by chromosomal location (from chr 1 to chr X) and signal-to-noise ratio (SNR). Gene Name Description SNR Score Feature P value FDR(BH) Q Value * CCL5 chemokine (C-C motif) ligand 5 1.395 0.002 0.036 0.025 * CCR7 chemokine (C-C motif) receptor 7 1.362 0.002 0.036 0.025 * CD79B CD79B antigen (immunoglobulin-associated beta) 1.248 0.002 0.036 0.025 * CCL13 chemokine (C-C motif) ligand 13 1.003 0.002 0.036 0.025 * CCL2 chemokine (C-C motif) ligand 2 0.737 0.004 0.056 0.037 * CCL8 chemokine (C-C motif) ligand 8 0.724 0.002 0.036 0.025 * CCL18 chemokine (C-C motif) ligand 18 (pulmonary and activ 0.703 0.002 0.036 0.025 * CCL23 chemokine (C-C motif) ligand 23 0.701 0.002 0.036 0.025 * CCR6 chemokine (C-C motif) receptor 6 0.715 0.002 0.036 0.025 PTPN7 protein tyrosine phosphatase, non-receptor type 7 1.861 0.002 0.036 0.025 CD3Z CD3Z antigen, zeta polypeptide (TiT3 complex) 1.811 0.002 0.036 0.025 CD48 CD48 antigen (B-cell membrane protein) 1.809 0.002 0.036 0.025 IL10RA interleukin 10 receptor, alpha 1.806 0.002 0.036 0.025 SELL selectin L (lymphocyte adhesion molecule 1) 1.773 0.002 0.036 0.025 LCK lymphocyte-specific protein tyrosine kinase 1.745 0.002 0.036 0.025 -

Neuromedins U and S Involvement in the Regulation of the Hypothalamo–Pituitary–Adrenal Axis
REVIEW ARTICLE published: 05 December 2012 doi: 10.3389/fendo.2012.00156 Neuromedins U and S involvement in the regulation of the hypothalamo–pituitary–adrenal axis Ludwik K. Malendowicz*, Agnieszka Ziolkowska and Marcin Rucinski Department of Histology and Embryology, Poznan University of Medical Sciences, Poznan, Poland Edited by: We reviewed neuromedin U (NMU) and neuromedin S (NMS) involvement in the regulation Hubert Vaudry, University of Rouen, of the hypothalamo–pituitary–adrenal (HPA) axis function. NMU and NMS are structurally France related and highly conserved neuropeptides. They exert biological effects via two GPCR Reviewed by: receptors designated as NMUR1 and NMUR2 which show differential expression. NMUR1 James A. Carr, Texas Tech University, USA is expressed predominantly at the periphery, while NMUR2 in the central nervous system. Gábor B. Makara, Hungarian Elements of the NMU/NMS and their receptors network are also expressed in the HPA Academy of Sciences, Hungary axis and progress in molecular biology techniques provided new information on their *Correspondence: actions within this system. Several lines of evidence suggest that within the HPA axis Ludwik K. Malendowicz, NMU and NMS act at both hypothalamic and adrenal levels. Moreover, new data suggest Department of Histology and Embryology, Poznan University of that NMU and NMS are involved in central and peripheral control of the stress response. Medical Sciences, 6 Swie¸cickiSt., 60-781 Poznan, Poland. Keywords: neuromedin U, neuromedin S, hypothalamus, pituitary, adrenal e-mail: [email protected] INTRODUCTION Identification of specific NMU receptors (NMUR1 and In search for new biologically active peptides, the group of NMUR2) and its anorexigenic action have enhanced interest in Minamino, Kangawa, and Matsuo in the 1980s isolated numerous physiological role of NMU and NMS (Howard et al., 2000; Ida small neuropeptides from porcine spinal cord. -

Oncogenic Features of Neuromedin U in Breast Cancer Are Associated with NMUR2 Expression Involving Crosstalk with Members of the WNT Signaling Pathway
www.impactjournals.com/oncotarget/ Oncotarget, 2017, Vol. 8, (No. 22), pp: 36246-36265 Research Paper Oncogenic features of neuromedin U in breast cancer are associated with NMUR2 expression involving crosstalk with members of the WNT signaling pathway Stefan Garczyk1, Natalie Klotz1, Sabrina Szczepanski1, Bernd Denecke2, Wiebke Antonopoulos1, Saskia von Stillfried1, Ruth Knüchel1, Michael Rose1,* and Edgar Dahl1,* 1Molecular Oncology Group, Institute of Pathology, Medical Faculty of the RWTH Aachen University, D-52074 Aachen, Germany 2IZKF Aachen, Medical Faculty of the RWTH Aachen University, D-52074 Aachen, Germany *Equal co-senior authors Correspondence to: Stefan Garczyk, email: [email protected] Keywords: breast cancer, metastasis, neuromedin U, neuromedin U receptor 2, WNT signaling Received: December 20, 2016 Accepted: February 07, 2017 Published: March 11, 2017 Copyright: Garczyk et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Neuromedin U (NMU) has been shown driving the progression of various tumor entities, including breast cancer. However, the expression pattern of NMU and its receptors in breast cancer tissues as well as systematic insight into mechanisms and downstream targets of the NMU-driven signaling pathways are still elusive. Here, NMU expression was found up-regulated in all breast cancer subtypes when compared to healthy breast tissue. Using an in silico dataset comprising 1,195 samples, high NMU expression was identified as an indicator of poor outcome in breast tumors showing strong NMUR2 expression. Next, the biological impact of NMU on breast cancer cells in relation to NMUR2 expression was analyzed. -

Cat. NMUR12-S Rabbit Anti-Human NMUR1 Antiserum # 2 SIZE: 100 Ul
Product Specification Sheet NMU Receptor 1 (NMUR1) Antibodies Cat. NMUR12-S Rabbit Anti-Human NMUR1 antiserum # 2 SIZE: 100 ul Cat. NMUR12-A Rabbit Anti-Human NMUR1 IgG # 2(aff pure) SIZE: 100 ug Cat. NMUR12-P Human NMUR1 Control/blocking peptide # 2 SIZE: 100 ug Form & Storage of Antibodies/Peptide Control The neuromedins (Nm) are a family of bioactive peptides best known for their roles in smooth muscle contraction. Nm family of Antiserum (unpurified) bioactive peptides include: Bombesin-like (NmB, NmC), kassinin- 100ul solution lyophilized powder like (NmL and NmK or neurokinins A and B), neurotensin-like Supplied in Buffer: 0.05% azide (NmN), and neuromedin U (NmU; for its ability to stimulate uterine Reconstitute powder in 100 ul PBS muscle contraction). Besides its roles in smooth muscle contraction (human ileum, urinary bladder, rat stomach etc), NmU has also been Affinity pure IgG implicated in hypertension, blood flow in intestine, and 100 ug/100ul solution lyophilized powder neurotransmission. Recently two structurally related, orphan G- Supplied in Buffer: PBS+0.1% BSA protein coupled receptors, termed NMUR1 (GPCR66/FM- Reconstitute powder in PBS at 1mg/ml 3/SNORF62) and NMUR2 (TGR-1/FM-4/SNORF72), have been identified as cognate receptors of NmU. NMU receptors display a Control/blocking peptide typical 7 TM domains with extracellular N-terminus and intracellular 100 ug/100 ul solution lyophilized powder C-terminus. Supplied in Buffer: PBS pH 7.5, Reconstitute powder in PBS at 1 mg/ml. The human NMUR1 (rat 402 aa; mouse, 405 aa; human 403 aa, chromosome 2; 70-80% interspecies homology) orphan GPCR66 Storage was originally identified by similarity to mouse orthologue FM-3. -

Ligand-Specific Signaling Profiles and Resensitization Mechanisms of the Neuromedin U2 Receptor
1521-0111/94/1/674–688$35.00 https://doi.org/10.1124/mol.117.111070 MOLECULAR PHARMACOLOGY Mol Pharmacol 94:674–688, July 2018 Copyright ª 2018 by The American Society for Pharmacology and Experimental Therapeutics Ligand-Specific Signaling Profiles and Resensitization Mechanisms of the Neuromedin U2 Receptor Khaled Alhosaini,1 Omar Bahattab,2 Heider Qassam, R. A. John Challiss, and Gary B. Willars Department of Molecular and Cell Biology, University of Leicester, Leicester, United Kingdom Received November 8, 2017; accepted April 24, 2018 Downloaded from ABSTRACT The structurally related, but distinct neuropeptides, neuromedin resensitization is faster following NmU compared with NmS U (NmU) and neuromedin S (NmS) are ligands of two G protein- exposure, but is similar if endothelin-converting enzyme-1 coupled NmU receptors (NMU1 and NMU2). Hypothalamic activity is inhibited or knocked down. Although acute activation NMU2 regulates feeding behavior and energy expenditure and of extracellular signal-regulated kinase (ERK) is also similar, has therapeutic potential as an anti-obesity target, making an activation by NMU2 is longer lasting if NmS is the ligand. understanding of its signaling and regulation of particular in- Furthermore, when cells are briefly challenged before removal molpharm.aspetjournals.org terest. NMU2 binds both NmU and NmS with high affinity, of free, but not receptor-bound ligand, activation of ERK and p38 resulting in receptor-ligand co-internalization. We have investi- mitogen-activated protein kinase by NmS is more sustained. gated whether receptor trafficking events post-internalization However, only NmU responses are potentiated and extended by are biased by the ligand bound and can therefore influence endothelin-converting enzyme-1 inhibition. -

Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Amy Sue Bogard University of Tennessee Health Science Center
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 12-2013 Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Amy Sue Bogard University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Medical Cell Biology Commons, and the Medical Molecular Biology Commons Recommended Citation Bogard, Amy Sue , "Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells" (2013). Theses and Dissertations (ETD). Paper 330. http://dx.doi.org/10.21007/etd.cghs.2013.0029. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Document Type Dissertation Degree Name Doctor of Philosophy (PhD) Program Biomedical Sciences Track Molecular Therapeutics and Cell Signaling Research Advisor Rennolds Ostrom, Ph.D. Committee Elizabeth Fitzpatrick, Ph.D. Edwards Park, Ph.D. Steven Tavalin, Ph.D. Christopher Waters, Ph.D. DOI 10.21007/etd.cghs.2013.0029 Comments Six month embargo expired June 2014 This dissertation is available at UTHSC Digital Commons: https://dc.uthsc.edu/dissertations/330 Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells A Dissertation Presented for The Graduate Studies Council The University of Tennessee Health Science Center In Partial Fulfillment Of the Requirements for the Degree Doctor of Philosophy From The University of Tennessee By Amy Sue Bogard December 2013 Copyright © 2013 by Amy Sue Bogard. -

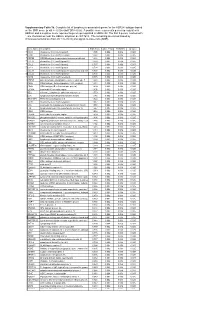
Neuromedin U Receptor 1 Expression in the Rat Endocrine Pancreas And
951-955 5/10/06 14:21 Page 951 INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 18: 951-955, 2006 951 Neuromedin U receptor 1 expression in the rat endocrine pancreas and evidence suggesting neuromedin U suppressive effect on insulin secretion from isolated rat pancreatic islets PRZEMYSLAW KACZMAREK1, LUDWIK K. MALENDOWICZ2, EWA PRUSZYNSKA-OSZMALEK1, TATIANA WOJCIECHOWICZ1, DAWID SZCZEPANKIEWICZ1, TOMASZ SZKUDELSKI1 and KRZYSZTOF W. NOWAK1 1Department of Animal Physiology and Biochemistry, August Cieszkowski University of Agriculture, 2Department of Histology and Embryology, Karol Marcinkowski University of Medical Sciences, Poznan, Poland Received May 25, 2006; Accepted July 19, 2006 Abstract. Neuromedin U (NmU) is a regulatory peptide are similar to other known neuromedins. They are commonly found in significant concentrations in both the brain and gut divided into four groups: bombesin-like (NmB, NmC), of the rat and is named according to its ability to powerfully kassinin-like (NmL/neurokinin A, NmK/neurokinin B), contract the uterus. Two types of NmU receptors were neurotensin-like (NmN) and neuromedin U (NmU). NmU was recently identified and subsequent studies evidenced NmU first isolated from extracts of porcine spinal cord over 15 involvement in the regulation of energy homeostasis. Such a years ago and was named for its uterine contractile activity role of neuromedin U suggests that a polypeptide may also (1,2). Two molecular forms of peptides exerting full biological be involved in the regulation of adipoinsular axis function. activity were identified: neuromedin U-25 (NmU-25) and its Therefore in the present study we examined the expression of cleavage product, neuromedin U-8 (NmU-8). Neuromedin U NmU receptors in pancreatic islets using RT-PCR and was subsequently isolated from a broad range of species Western blotting analysis.