Development of an Expression Vector to Overexpress Or Downregulate Genes in Curvularia Protuberata
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Therapy Glossary of Terms
GENE THERAPY GLOSSARY OF TERMS A • Phase 3: A phase of research to describe clinical trials • Allele: one of two or more alternative forms of a gene that that gather more information about a drug’s safety and arise by mutation and are found at the same place on a effectiveness by studying different populations and chromosome. different dosages and by using the drug in combination • Adeno-Associated Virus: A single stranded DNA virus that has with other drugs. These studies typically involve more not been found to cause disease in humans. This type of virus participants.7 is the most frequently used in gene therapy.1 • Phase 4: A phase of research to describe clinical trials • Adenovirus: A member of a family of viruses that can cause occurring after FDA has approved a drug for marketing. infections in the respiratory tract, eye, and gastrointestinal They include post market requirement and commitment tract. studies that are required of or agreed to by the study • Adeno-Associated Virus Vector: Adeno viruses used as sponsor. These trials gather additional information about a vehicles for genes, whose core genetic material has been drug’s safety, efficacy, or optimal use.8 removed and replaced by the FVIII- or FIX-gene • Codon: a sequence of three nucleotides in DNA or RNA • Amino Acids: building block of a protein that gives instructions to add a specific amino acid to an • Antibody: a protein produced by immune cells called B-cells elongating protein in response to a foreign molecule; acts by binding to the • CRISPR: a family of DNA sequences that can be cleaved by molecule and often making it inactive or targeting it for specific enzymes, and therefore serve as a guide to cut out destruction and insert genes. -

Lecture 3 Types, Biology and Salient Features of Vectors in Recombinant
NPTEL – Bio Technology – Genetic Engineering & Applications MODULE 1- LECTURE 3 TYPES, BIOLOGY AND SALIENT FEATURES OF VECTORS IN RECOMBINANT DNA TECHNOLOGY – PLASMID 1-3.1 Introduction: DNA molecule used for carrying an exogenous DNA into a host organism and facilitates stable integration and replication inside the host system is termed as Vector. Molecular cloning involves series of sequential steps which includes restriction digestion of DNA fragments both target DNA and vector, ligation of the target DNA with the vector and introduction into a host organism for multiplication. Then the fragments resulted after digestion with restriction enzymes are ligated to other DNA molecules that serve as vectors. In general, vectors should have following characteristics: • Capable of replicating inside the host. • Have compatible restriction site for insertion of DNA molecule (insert). • Capable of autonomous replication inside the host (ori site). • Smaller in size and able to incorporate larger insert size. • Have a selectable marker for screening of recombinant organism. 1-3.2 Plasmids: • Plasmids are naturally occurring extra chromosomal double-stranded circular DNA molecules which can autonomously replicate inside bacterial cells. Plasmids range in size from about 1.0 kb to over 250 kb. • Plasmids encode only few proteins required for their own replication (replication proteins) and these proteins encoding genes are located very close to the ori. All the other proteins required for replication, e.g. DNA polymerases, DNA ligase, helicase, etc., are provided by the host cell.Thus, only a small region surrounding Joint initiative of IITs and IISc – Funded by MHRD Page 28 of 84 NPTEL – Bio Technology – Genetic Engineering & Applications the ori site is required for replication. -

Molecular Biology and Applied Genetics
MOLECULAR BIOLOGY AND APPLIED GENETICS FOR Medical Laboratory Technology Students Upgraded Lecture Note Series Mohammed Awole Adem Jimma University MOLECULAR BIOLOGY AND APPLIED GENETICS For Medical Laboratory Technician Students Lecture Note Series Mohammed Awole Adem Upgraded - 2006 In collaboration with The Carter Center (EPHTI) and The Federal Democratic Republic of Ethiopia Ministry of Education and Ministry of Health Jimma University PREFACE The problem faced today in the learning and teaching of Applied Genetics and Molecular Biology for laboratory technologists in universities, colleges andhealth institutions primarily from the unavailability of textbooks that focus on the needs of Ethiopian students. This lecture note has been prepared with the primary aim of alleviating the problems encountered in the teaching of Medical Applied Genetics and Molecular Biology course and in minimizing discrepancies prevailing among the different teaching and training health institutions. It can also be used in teaching any introductory course on medical Applied Genetics and Molecular Biology and as a reference material. This lecture note is specifically designed for medical laboratory technologists, and includes only those areas of molecular cell biology and Applied Genetics relevant to degree-level understanding of modern laboratory technology. Since genetics is prerequisite course to molecular biology, the lecture note starts with Genetics i followed by Molecular Biology. It provides students with molecular background to enable them to understand and critically analyze recent advances in laboratory sciences. Finally, it contains a glossary, which summarizes important terminologies used in the text. Each chapter begins by specific learning objectives and at the end of each chapter review questions are also included. -

Investigations on the Occurrence of Fungal Root Endophytes and an Associated Mycovirus in Context with Apple Replant Disease
Investigations on the occurrence of fungal root endophytes and an associated mycovirus in context with apple replant disease Von der Naturwissenschaftlichen Fakultät der Gottfried Wilhelm Leibniz Universität Hannover zur Erlangung des Grades Doktorin der Gartenbauwissenschaften (Dr. rer. hort.) genehmigte Dissertation von Gösta Carolin Dorette Popp, M. Sc. 2020 Referent: Prof. Dr. Edgar Maiss Korreferentin: Prof. Dr. Traud Winkelmann Tag der Promotion: 12.08.2020 Abstract I Abstract Apple replant disease (ARD) negatively affects the production in nurseries and orchards worldwide. A biotic cause of the disease is most likely since soil disinfection can restore plant growth. Fungi have appeared to contribute to the complex of biotic factors, but up to now the actual cause of the disease remains unknown. Further, environmentally friendly and practically applicable mitigation strategies are missing. Fungal root endophytes were isolated in two central experiments of the ORDIAmur consortium. Dark septate endophytes (Leptodontidium spp.) were frequently isolated from apple roots. An abundant occurrence of Nectriaceae fungi (Dactylonectria torresensis and Ilyonectria robusta) was found in ARD roots. Reference sites displayed a different characteristic fungal community. In roots grown in irradiated soil, a reduction of the number of isolated fungi and a changed composition of the fungal community was found. To investigate the effect of fungal endophytes on apple plants a quick and soil-free bio test in Petri dishes was developed using perlite. Inoculated fungi isolated from ARD roots induced neutral (Plectosphaerella, Pleotrichocladium, and Zalerion) to negative (Cadophora, Calonectria, Dactylonectria, Ilyonectria, and Leptosphaeria) plant reactions. After re-isolation, most of the Nectriaceae isolates were confirmed as pathogens. Microscopic analyses of ARD-affected roots revealed necroses caused by an unknown fungus that forms cauliflower-like (CF) structures in diseased cortex cells. -

Regulation of Transgene Expression in Genetic Immunization
Brazilian Journal of Medical and Biological Research (1999) 32: 155-162 Regulation of transgene expression 155 ISSN 0100-879X Regulation of transgene expression in genetic immunization J.S. Harms1, 1Department of Animal Health and Biomedical Sciences, University of Wisconsin, S.C. Oliveira2 Madison, WI, USA and G.A. Splitter1 2Departamento de Bioquímica e Imunologia, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brasil Abstract Correspondence The use of mammalian gene expression vectors has become increas- Key words J.S. Harms ingly important for genetic immunization and gene therapy as well as · DNA vaccine AHABS basic research. Essential for the success of these vectors in genetic · Gene therapy 1655 Linden Drive · immunization is the proper choice of a promoter linked to the antigen Regulation Madison, WI 53706 · Transcription of interest. Many genetic immunization vectors use promoter elements USA · Transgene Fax: +1-608-262-7420 from pathogenic viruses including SV40 and CMV. Lymphokines E-mail: [email protected] produced by the immune response to proteins expressed by these vectors could inhibit further transcription initiation by viral promot- Research supported by the Robert ers. Our objective was to determine the effect of IFN-g on transgene Draper Technology Innovation Fund expression driven by viral SV40 or CMV promoter/enhancer and the (No. 135-0546) of the University mammalian promoter/enhancer for the major histocompatibility com- of Wisconsin, Madison. plex class I (MHC I) gene. We transfected the luciferase gene driven Presented at the International by these three promoters into 14 cell lines of many tissues and several Symposium “The Third Revolution species. -

RNA Viruses As Tools in Gene Therapy and Vaccine Development
G C A T T A C G G C A T genes Review RNA Viruses as Tools in Gene Therapy and Vaccine Development Kenneth Lundstrom PanTherapeutics, Rte de Lavaux 49, CH1095 Lutry, Switzerland; [email protected]; Tel.: +41-79-776-6351 Received: 31 January 2019; Accepted: 21 February 2019; Published: 1 March 2019 Abstract: RNA viruses have been subjected to substantial engineering efforts to support gene therapy applications and vaccine development. Typically, retroviruses, lentiviruses, alphaviruses, flaviviruses rhabdoviruses, measles viruses, Newcastle disease viruses, and picornaviruses have been employed as expression vectors for treatment of various diseases including different types of cancers, hemophilia, and infectious diseases. Moreover, vaccination with viral vectors has evaluated immunogenicity against infectious agents and protection against challenges with pathogenic organisms. Several preclinical studies in animal models have confirmed both immune responses and protection against lethal challenges. Similarly, administration of RNA viral vectors in animals implanted with tumor xenografts resulted in tumor regression and prolonged survival, and in some cases complete tumor clearance. Based on preclinical results, clinical trials have been conducted to establish the safety of RNA virus delivery. Moreover, stem cell-based lentiviral therapy provided life-long production of factor VIII potentially generating a cure for hemophilia A. Several clinical trials on cancer patients have generated anti-tumor activity, prolonged survival, and -

Clone the Unclonable – Vectors and Cells to Capture and Express Problematic Dnas
Clone the Unclonable – Vectors and Cells to Capture and Express Problematic DNAs Ron Godiska, Ph.D. May, 2016 Agenda Improve Cloning and Expression of Problematic DNA • What makes DNA difficult to clone or express? • Circular vectors for difficult DNAs • Linear vectors for “impossible” DNAs • Enzyme-free cloning and compatible protein expression vectors • E. coli strains for toxic recombinant proteins or unstable DNA What is Unclonable DNA? Characteristics of Difficult DNA • Toxic coding sequences • Promoters • A-T Rich DNA • Large fragments (>10 kb) • Trace amounts Which types of difficult DNA have you worked with in the lab? Common Vector Traits Cause Cloning Problems Vector driven transcription into the insert – Deleterious expression – Destabilize 2o structure Insert driven transcription out into the vector – False negatives (blue or lethal) – Transcriptional interference pUC19 – Replication interference High copy number – Increased effect of the above issues – Active origin – Many copies/cell Supercoiling – Deletion of secondary structures CloneSmart® Technology “Silencing” DNA Inserts to Improve Cloning • No Transcriptional Interference – Stably maintain “unclonable” DNA • No blue/white screening – No false positives/negatives • Minimal vector size • Variable copy number • Variety of drug selection markers • Very low empty-vector background U.S. Patent 6,709,861 pSMART Vectors Clone Toxic Inserts and Promoters Test DNA Fragments: • RNase coding sequence (350 bp; no promoter) • Phage Lambda PR Promoter (400 bp) pSMART Vectors cDNA Clones are More Stable Deletion Rates: <1% in pSMART HC Kan vs. 32% in pUC % Deleted Clones Insert Size (kb) • Teleost fish cDNA library • Clones were grown in liquid medium, diluted, and regrown • Each final culture was diluted, plated, and grown up into individual colonies • Plasmid DNA was isolated from colonies for each clone and analyzed - Oleksiak M.F., Crawford D.L., 2001. -
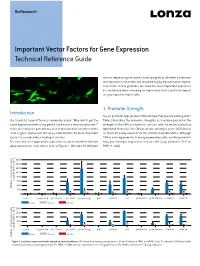
Important Vector Factors for Gene Expression Technical Reference Guide
BioResearch Important Vector Factors for Gene Expression Technical Reference Guide vectors expressing the same luciferase gene in different backbones and expression cassettes and obtained highly discriminative expres- sion levels. In this guideline, we show the most important aspects to be considered when choosing an expression vector and their impact on your experimental results. 1. Promoter Strength Introduction Is your promoter appropriate for the cell type that you are working with? Our Scientific Support Team is commonly asked: “Why don’t I get the Table 1 describes the promoter strengths as a relative percent of the same expression level of my gene if I use various vector backbones?” strength of the CMV promoter for various cells for which Lonza has There are many components to a vector that can have an effect on the Optimized Protocols. The CMV promoter activity is set to 100% based level of gene expression. Below you will find the 10 most important on their CAT assay values from the referenced publications. Although factors to consider when looking at vectors. CMV is a strong promoter in many mammalian cells, another promoter The selection of an appropriate expression vector is crucial for efficient may give stronger expression in your cells (e.g., promoter SV40 in gene expression. Just take a look at Figure 1. We tried 10 different BHK-21 cells). n 1600 1400 1200 1000 800 [% of pGL3-CMV at 24 h] 24 at pGL3-CMV [% of 600 400 Relative luciferase-expressio Relative 200 0 Control Program Only pGL3-Control pGL3-CMV pmax Cloning™ pSG5 pcDNA3.1 pFP pIRES-MCS B pcDNA4 HisMax pCMVbeta pCMV-TNT 4 hours 24 hours 48 hours n 2000 1500 1000 500 0 Control Program Only pGL3-Control pGL3-CMV pmax Cloning™ pSG5 pcDNA3.1 pFP pIRES-MCS B pcDNA4 HisMax pCMVbeta pCMV-TNT [% of pGL3-CMV at 24 h] 24 at pGL3-CMV [% of 4 hours 24 hours 48 hours Relative luciferase-expressio Relative Figure 1: Luciferase expression levels depend on vector backbones. -

Cloning Technologies for Protein Expression and Purification
Cloning technologies for protein expression and purification James L Hartley Detailed knowledge of the biochemistry and structure of expedite the manipulations that are often required to individual proteins is fundamental to biomedical research. To obtain pure, active recombinant proteins. further our understanding, however, proteins need to be purified in sufficient quantities, usually from recombinant sources. Cloning for protein expression and Although the sequences of genomes are now produced in purification automated factories purified proteins are not, because their The required amount, purity, homogeneity, and activity behavior is much more variable. The construction of plasmids of the protein of interest usually determine the choice of and viruses to overexpress proteins for their purification is often expression context, vector and host. Usually the first tedious. Alternatives to traditional methods that are faster, easier consideration is what host cell to use. Expression in and more flexible are needed and are becoming available. Escherichia coli is fast, inexpensive and scaleable, and minimal post-translational modifications make proteins Addresses Protein Expression Laboratory, Research Technology Program, purified from E. coli relatively homogeneous and highly SAIC-Frederick, Inc., NCI-Frederick, Frederick, Maryland 21702, USA desirable for structural studies. However, in our experi- ence most eukaryotic proteins larger than 30 kDa are Corresponding author: Hartley, James L ([email protected]) unlikely to be properly folded when expressed in E. coli (JL Hartley et al., unpublished). Expression in mamma- Current Opinion in Biotechnology 2006, 17:359–366 lian cells is best for activity and native structure (includ- ing post-translational modifications), but yields are much This review comes from a themed issue on lower and costs are high. -

Australia Biodiversity of Biodiversity Taxonomy and and Taxonomy Plant Pathogenic Fungi Fungi Plant Pathogenic
Taxonomy and biodiversity of plant pathogenic fungi from Australia Yu Pei Tan 2019 Tan Pei Yu Australia and biodiversity of plant pathogenic fungi from Taxonomy Taxonomy and biodiversity of plant pathogenic fungi from Australia Australia Bipolaris Botryosphaeriaceae Yu Pei Tan Curvularia Diaporthe Taxonomy and biodiversity of plant pathogenic fungi from Australia Yu Pei Tan Yu Pei Tan Taxonomy and biodiversity of plant pathogenic fungi from Australia PhD thesis, Utrecht University, Utrecht, The Netherlands (2019) ISBN: 978-90-393-7126-8 Cover and invitation design: Ms Manon Verweij and Ms Yu Pei Tan Layout and design: Ms Manon Verweij Printing: Gildeprint The research described in this thesis was conducted at the Department of Agriculture and Fisheries, Ecosciences Precinct, 41 Boggo Road, Dutton Park, Queensland, 4102, Australia. Copyright © 2019 by Yu Pei Tan ([email protected]) All rights reserved. No parts of this thesis may be reproduced, stored in a retrieval system or transmitted in any other forms by any means, without the permission of the author, or when appropriate of the publisher of the represented published articles. Front and back cover: Spatial records of Bipolaris, Curvularia, Diaporthe and Botryosphaeriaceae across the continent of Australia, sourced from the Atlas of Living Australia (http://www.ala. org.au). Accessed 12 March 2019. Taxonomy and biodiversity of plant pathogenic fungi from Australia Taxonomie en biodiversiteit van plantpathogene schimmels van Australië (met een samenvatting in het Nederlands) Proefschrift ter verkrijging van de graad van doctor aan de Universiteit Utrecht op gezag van de rector magnificus, prof. dr. H.R.B.M. Kummeling, ingevolge het besluit van het college voor promoties in het openbaar te verdedigen op donderdag 9 mei 2019 des ochtends te 10.30 uur door Yu Pei Tan geboren op 16 december 1980 te Singapore, Singapore Promotor: Prof. -

Complete Control Retroviral Inducible Mammalian Expression System
Complete Control Retroviral Inducible Mammalian Expression System Instruction Manual Catalog #217564 (pFB-ERV), #240028 (pCFB-EGSH) Revision D.0 For Research Use Only. Not for use in diagnostic procedures. 217564-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement of this product. No other warranties of any kind, express or implied, including without limitation, implied warranties of merchantability or fitness for a particular purpose, are provided by Agilent. Agilent shall have no liability for any direct, indirect, consequential, or incidental damages arising out of the use, the results of use, or the inability to use this product. ORDERING INFORMATION AND TECHNICAL SERVICES Email [email protected] World Wide Web www.genomics.agilent.com Telephone Location Telephone United States and Canada 800 227 9770 Austria 01 25125 6800 Benelux 02 404 92 22 Denmark 45 70 13 00 30 Finland 010 802 220 France 0810 446 446 Germany 0800 603 1000 Italy 800 012575 Netherlands 020 547 2600 Spain 901 11 68 90 Sweden 08 506 4 8960 Switzerland 0848 8035 60 UK/Ireland 0845 712 5292 All Other Countries Please visit www.agilent.com/genomics/contactus Complete Control Retroviral Inducible Mammalian Expression System Materials Provided .............................................................................................................................. 1 Storage Conditions .............................................................................................................................. 1 Additional Materials -

An Episomal Expression Vector for Screening Mutant Gene Libraries in Pichia Pastoris
Plasmid 54 (2005) 80–85 www.elsevier.com/locate/yplas Short communication An episomal expression vector for screening mutant gene libraries in Pichia pastoris Charles C. Lee*, Tina G. Williams, Dominic W.S. Wong, George H. Robertson USDA-ARS-WRRC, 800 Buchanan St., Albany, CA 94710, USA Received 7 October 2004, revised 6 December 2004 Available online 20 January 2005 Communicated by Manuel Espinosa Abstract Screening mutant gene libraries for isolating improved enzyme variants is a powerful technique that benefits from effective and reliable biological expression systems. Pichia pastoris is a very useful organism to express proteins that are inactive in other hosts such as Escherichia coli and Saccharomyces cerevisiae. However, most P. pastoris expression plasmids are designed to integrate into the host chromosome and hence are not as amenable to high-throughput screen- ing projects. We have designed a P. pastoris expression vector, pBGP1, incorporating an autonomous replication sequence that allows the plasmid to exist as an episomal element. This vector contains the a-factor signal sequence to direct secretion of the mutant enzymes. Expression of the genes is driven by the constitutive GAP promoter, thus eliminating the need for timed or cell density-specific inductions. The pBGP1 plasmid was used to screen a xylanase gene library to isolate higher activity mutants. Published by Elsevier Inc. Keywords: Pichia pastoris; Episomal expression vector 1. Introduction formed with libraries of randomly mutated genes encoding the enzyme of interest (Cohen et al., There are many efforts directed at improving 2001). Clones that demonstrate an increased activ- enzymes involved in industrial processes in order ity relative to wild type can be isolated and further to increase cost and energy efficiency.